Abstract.
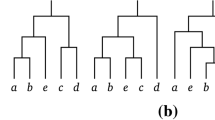
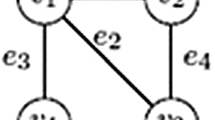
The graph-theoretic operation of rooted subtree prune and regraft is increasingly being used as a tool for understanding and modelling reticulation events in evolutionary biology. In this paper, we show that computing the rooted subtree prune and regraft distance between two rooted binary phylogenetic trees on the same label set is NP-hard. This resolves a longstanding open problem. Furthermore, we show that this distance is fixed parameter tractable when parameterised by the distance between the two trees.
Similar content being viewed by others
Author information
Authors and Affiliations
Corresponding author
Additional information
Received March 16, 2004
Rights and permissions
About this article
Cite this article
Bordewich, M., Semple, C. On the Computational Complexity of the Rooted Subtree Prune and Regraft Distance. Ann. Comb. 8, 409–423 (2005). https://doi.org/10.1007/s00026-004-0229-z
Issue Date:
DOI: https://doi.org/10.1007/s00026-004-0229-z