Abstract
Key message
Analysis of 387 sugarcane clones using Bru 1 diagnostic markers revealed two possible sources of Bru 1 in Chinese cultivars: one from Saccharum spontaneum and another from Saccharum robustum of New Guinea.
Abstract
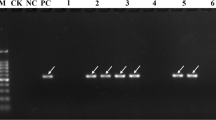
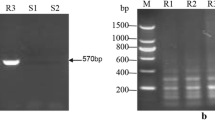
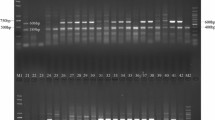
Sugarcane brown rust (SBR) is an important fungal disease in many sugarcane production areas around the world, and can cause considerable yield losses in susceptible sugarcane cultivars. One major SBR resistance gene, named Bru1, initially identified from cultivar R570, was shown to be a major SBR resistance source in most of the sugarcane producing areas of the world. In this study, by using the two Bru1-associated markers, R12H16 and 9O20-F4, we surveyed the presence of Bru1 in a Chinese sugarcane germplasm collection of 387 clones, consisting of 228 hybrid cultivars bred by different Chinese sugarcane breeding establishments, 54 exotic hybrid cultivars introduced from other countries and 105 clones of sugarcane ancestral species. The Bru1-bearing haplotype was detected in 43.4% of Chinese sugarcane cultivars, 20.4% of exotic hybrid cultivars, and only 3.8% of ancestral species. Among the 33 Chinese cultivars for which phenotypes of resistance to SBR were available, Bru1 was present in 69.2% (18/26) of the resistant clones. Analyses of the allelic sequence variations of R12H16 and 9O20-F4 suggested two possible sources of Bru1 in Chinese cultivars: one from S. spontaneum and another from S. robustum of New Guinea. In addition, we developed an improved Bru1 diagnostic marker, 9O20-F4-HaeIII, which can eliminate all the false results of 9O20-F4-RsaI observed among S. spontaneum, as well as a new dominant Bru1 diagnostic marker, R12E03-2, from the BAC ShCIR12E03. Our results provide valuable information for further efforts of breeding SBR-resistant varieties, searching new SBR resistance sources and cloning of Bru1 in sugarcane.



Similar content being viewed by others
References
Amalraj VA, Balasundaram N (2006) On the taxonomy of the members of ‘Saccharum complex’. Genet Resour Crop Evol 53:35–41
Asnaghi C, Paulet F, Kaye C, Grivet L, Deu M, Glaszmann JC, D’Hont A (2000) Application of synteny across Poaceae to determine the map location of a sugarcane rust resistance gene. Theor Appl Genet 101:962–969
Asnaghi C, D’hont A, Glaszmann J, Rott P (2001) Resistance of sugarcane cultivar R 570 to Puccinia melanocephala isolates from different geographic locations. Plant Dis 85:282–286
Asnaghi C, Roques D, Ruffel S, Kaye C, Hoarau JY, Telismart H, Girard JC, Raboin LM, Risterucci AM, Grivet L, D’Hont A (2004) Targeted mapping of a sugarcane rust resistance gene (Bru1) using bulked segregant analysis and AFLP markers. Theor Appl Genet 108:759–764
Bhatt SR, Gill SS (1985) The implications of 2n egg gametes in nobilization and breeding of sugarcane. Euphytica 34:377–384
Bremer G (1961a) Problems in breeding and cytology of sugarcane. I. A short history of sugarcane breeding—the original forms of Saccharum. Euphytica 10(1):59–78
Bremer G (1961b) Problems in breeding and cytology of sugarcane. II. The sugar cane breeding from a cytological view-point. Euphytica 10(2):121–258
Canilha L, Chandel AK, Milessi TSS, Antunes FAF, Freitas WLC, Felipe MGA, Silva SS (2012) Bioconversion of sugarcane biomass into ethanol: an overview about composition, pretreatment methods, detoxification of hydrolysates, enzymatic saccharification and ethanol fermentation. J Biomed Biotechnol 8:989572
Chandel AK, Silva SS, Carvalho W, Singh OV (2012) Sugarcane bagasse and leaves: foreseeable biomass of biofuel and bio-products. J Chem Technol Biotechnol 87:1–20
Comstock JC, Shine J Jr, Raid RN (1992) Effect of rust on sugarcane growth and biomass. Plant Dis 76:175–177
Comstock JC, Sood SG, Glynn NC, McKemy Shine JJJ, Castlebury LA (2008) First report of Puccinia kuehnii, causal agent of orange rust of sugarcane, in the United States and Western Hemisphere. Plant Dis 92:175
Costet L, Le Cunff L, Royaert S, Raboin LM, Hervouet C, Toubi L, Telismart H, Garsmeur O, Rousselle Y, Pauquet J, Nibouche S, Glaszmann JC, Hoarau JY, D’Hont A (2012) Haplotype structure around Bru1 reveals a narrow genetic basis for brown rust resistance in modern sugarcane cultivars. Theor Appl Genet 125(5):825–836
D’Hont A, Grivet L, Feldmann P, Rao PS, Berding N (1996) Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp) by molecular cytogenetics. Mol Gen Genet 250:405–413
D’Hont A, Ison D, Alix K, Roux C, Glaszmann JC (1998) Determination of basic chromosome numbers in the genus Saccharum by physical mapping of RNA genes. Genome 41:221–225
D’Hont A, Paulet F, Glaszmann JC (2002) Oligoclonal interspecific origin of ‘North Indian’ and ‘Chinese’ sugarcanes. Chromosome Res 10:253–262
Daniels J, Daniels CA (1975) Geographical, historical and cultural aspects of the origin of the Indian and Chinese sugarcanes S. barberi and S. sinense. Sugarcane Breed Newslett 36:4–23
Daniels J, Roach BT (1987) Taxonomy and evolution. In: Heinz DJ (ed) Sugarcane improvement through breeding. Elsevier Press, Amsterdam, pp 7–84
Daniels J, Smith P, Paton N, Williams CA (1975) The origin of the genus Saccharum. Sugarcane Breed Newsett 36:24–39
Daugrois JH, Grivet L, Roques D, Hoarau JY, Lombard H, Glaszmann JC, D’Hont A (1996) A putative major gene for rust resistance linked with a RFLP marker in sugarcane cultivar‘R570’. Theor Appl Genet 92:1059–1064
Dean JL, Tai PYP, Todd EH (1979) Sugarcane rust in Florida. Sugar J 42:10
D’Hont A, Lu YH, Feldmann P, Glaszmann JC (1993) Cytoplasmic diversity in sugarcane revealed by heterologous probes. Sugar Cane 1993(1):12–15
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Garsmeur O, Charron C, Bocs S, Jouffe V, Samain S, Couloux A, Droc G, Zini C, Glaszmann JC, Van Sluys MA, D’Hont A (2011) High homologous gene conservation despite extreme autopolyploid redundancy in sugarcane. New Phytol 189(2):629–642
Glaszmann JC, Lu YH, Lanaud C (1990) Variation of nuclear ribosomal DNA in sugarcane. J Genet Breed 44:191–198
Glynn NC, Laborde C, Davidson RW, Irey MS, Glaz B, D’Hont A, Comstock JC (2013) Utilization of a major brown rust resistance gene in sugarcane breeding. Mol Breed 31(2):323–331
Grivet L, Glaszmann JC, D’Hont A (2005) Molecular evidence of sugarcane evolution and domestication. In: Motley TJ, Zerega N, Cross H (eds) Darwin’s harvest: new approaches to the origins, evolution, and conservation of crops. Columbia University Press, New York, pp 49–66
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–99
Hoarau JY, Offmann B, D’Hont A, Risterucci AM, Roques D, Glaszmann JC, Grivet L (2001) Genetic dissection of a modern sugarcane cultivar (Saccharum spp.). I. Genome mapping with AFLP markers. Theor Appl Genet 103:84–97
Hoy JW, Hollier CA (2009) Effect of brown rust on yield of sugarcane in Louisiana. Plant Dis 93:1171–1174
Larkin MA, Blackshields G, Brown NP, Chenna R, Mc Gettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947
Le Cunff L, Garsmeur O, Raboin LM, Pauquet J, Telismart H, Selvi A, Grivet L, Philippe R, Begum D, Deu M, Costet L, Wing R, Glaszmann JC, D’Hont A (2008) Diploid/polyploid syntenic shuttle mapping and haplotype-specific chromosome walking toward a rust resistance gene (Bru1) in highly polyploid sugarcane (2n ~ 12x ~ 115). Genetics 180(1):649–660
Li WF, Wang XY, Huang YK, Zhang RY, Shan HL, Yin J, Luo ZM (2017) Molecular detection of Bru1 gene and identification of brown rust resistance in chinese sugarcane germplasm. Sugar Technol 19(2):183–190
Liu YQ, Li YP, Liang WH, Song QD, Qin XL, Ye L (2015) Current situation of sugar cane industry in the world. World Agriculture 2015(8):147–152
Lu YH, D’Hont A, Walker DIT, Rao PS, Feldmann P, Glaszmann JC (1994a) Relationships among ancestral species of sugarcane revealed with RFLP using single copy maize nuclear probes. Euphytica 78:7–18
Lu YH, D’Hont A, Paulet F, Grivet L, Arnaud M, Glaszmann JC (1994b) Molecular diversity and genome structure in modern sugarcane varieties. Euphytica 78:216–226
Luo J, Pan YB, Xu L, Zhang H, Yuan Z, Deng Z, Chen R, Que Y (2014) Cultivar evaluation and essential test locations identification for sugarcane breeding in China. Sci World J 2014:302753
Mukherjee SK (1957) Origin and distribution of Saccharum. Bot Gaz 119:55–61
Panje RR (1972) The role of Saccharum spontaneum in sugarcane breeding. Proc Int Soc Sugar Cane Technol 14:217–223
Panje RR, Babu CN (1960) Studies in Saccharum spontaneum: distribution and geographic association of chromo-some numbers. Cytologia (Tokyo) 25:152–172
Parco AS, Avellaneda MC, Hale AH, Hoy JW, Kimbeng CA, Pontif MJ, Gravois KA, Baisakh N (2014) Frequency and distribution of the brown rust resistance gene Bru1 and implications for the Louisiana sugarcane breeding programme. Plant Breed 133(5):654–659
Peixoto-Junior RF, Creste S, Landell MGA, Nunes DS, Sanguino A, Campos MF, Vencovsky R, Tambarussi EV, Figueira A (2014) Genetic diversity among Puccinia melanocephala isolates from Brazil assessed using simple sequence repeat markers. Genet Mol Res 13(3):7852–7863
Peng SG (1980) The general situation of evolution of sugarcane varieties at home and abroad. J South Agric 10:12–15
Peng SG (1996) Varietal evolution of sugarcane in Taiwan. Southwest China J Agric Sci 9(1):117–124
Piperidis G, Piperidis N, D’Hont A (2010) Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol Genet Genom 284(1):65–73
Pocovi MII, Rech GE, Collavino NG, Caruso GB, Rios R, Mariotti JA (2010) Molecular diversity of Puccinia melanocephala populations. J Phytopathol 158:769–775
Price S (1968) Cytology of Chinese and North Indian sugarcanes. Econ Bot 22:155–164
Purdy LH, Liu JL, Dean JL (1983) Sugarcane rust, a newly important disease. Plant Dis 67:1292–1295
Que Y, Xu L, Wu Q, Liu Y, Ling H, Liu Y, Zhang Y, Guo J, Su Y, Chen J, Wang S, Zhang C (2014) Genome sequencing of Sporisorium scitamineum provides insights into the pathogenic mechanisms of sugarcane smut. BMC Genom 15:996
Raboin LM, Oliveira KM, Lecunff L, Telismart H, Roques D, Butterfield M, Hoarau JY, D’Hont A (2006) Genetic mapping in sugarcane, a high polyploid, using bi-parental progeny: identification of a gene controlling stalk colour and a new rust resistance gene. Theor Appl Genet 112:1382–1391
Racedo J, Perera MF, Bertani R, Funes C, González V, Cuenya MI, D’Hont A, Welin B, Castagnaro AP (2013) Bru1 gene and potential alternative sources of resistance to sugarcane brown rust disease. Euphytica 191:429–436
Roach BT (1972) Nobilization of sugarcane. Proc Int Soc Sugar Cane Technol 14:206–216
Roach BT (1977) Utilization of Saccharum spontaneum in sugarcane breeding. Proc Int Soc Sugar Cane Technol 16:43–57
Ruan XY, Yan F, Sun CJ (1983) Occurrence of Puccinia erianthi on sugarcane in Yunnan province. Acta Mycologica Sinia 2:260–261
Sreenivasan TV, Ahloowalia BS, Heinz DJ (1987) Cytogenetics. In: Heinz DJ (ed) Sugarcane improvement through breeding. Elsevier, Amsterdam, pp 143–210
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Todd J, Glaz B, Burner D, Kimbeng C (2015) Historical use of cultivars as parents in Florida and Louisiana sugarcane breeding programs. Int Sch Res Not 2015:257417
Wang XY, Li WF, Huang YK, Lu X, Luo ZH, Yin J, Shan HL, Zhang RY (2013) Evaluation of sugarcane introgression lines for resistance to brown rust disease caused by Puccinia melanocephala. Trop Plant Pathol 38(2):097–101
Wei Q, Yang BL, Gao ZJ (2015) Analysis of current situation of sugar cane industry. J Agric Mech Res 4:247–254
Zhang XY (1996) Utilization of S. robustum descendants in sugarcane breeding. Sugarcane 3(1):14–18
Zhang YX (2003) Sugarcane breeding strategies in Taiwan. Sugarcane Canesugar 2003(1):22–25
Zhang Q, Qi YW, Zhang CM, Chen YS, Deng HH (2009) Pedigree analysis of genetic relationship among core parents of sugarcane in Mainland China. Guangdong Agric Sci 2009(10):44–48
Zhang J, Sharma A, Yu Q, Wang J, Li L, Zhu L, Zhang X, Chen Y, Ming R (2016) Comparative structural analysis of Bru1 region homeologs in Saccharum spontaneum and S. officinarum. BMC Genom 17:446
Zhou M (2013) Conventional sugarcane breeding in South Africa: progress and future prospects. Am J Plant Sci 4:189–196
Acknowledgements
This work was supported by a startup fund for distinguished scholars of Fujian Agriculture and Forestry University, and the Project of Education and Scientific Research of Young Teacher of Fujian (JA13102). The authors would like to thank Xin Lu for providing facilities and help in obtaining the leaf tissues from NNSGR.
Author information
Authors and Affiliations
Contributions
YL conceived and designed the experiments; HW, PC and YY conducted the experiments; HW and YL processed the data; YL wrote the manuscript; AD reviewed the manuscript; all authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the experiments presented in this publication comply with the current laws of China and France.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Communicated by Dr. Antonio Augusto Franco Garcia.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, HB., Chen, PH., Yang, YQ. et al. Molecular insights into the origin of the brown rust resistance gene Bru1 among Saccharum species. Theor Appl Genet 130, 2431–2443 (2017). https://doi.org/10.1007/s00122-017-2968-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2968-3