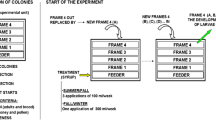
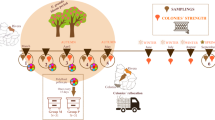
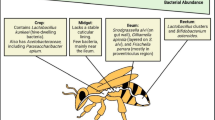
Abstract
Honeybees are important pollinators, having an essential role in the ecology of natural and agricultural environments. Honeybee colony losses episodes reported worldwide and have been associated with different pests and pathogens, pesticide exposure, and nutritional stress. This nutritional stress is related to the increase in monoculture areas which leads to a reduction of pollen availability and diversity. In this study, we examined whether nutritional stress affects honeybee gut microbiota, bee immunity, and infection by Nosema ceranae, under laboratory conditions. Consumption of Eucalyptus grandis pollen was used as a nutritionally poor-quality diet to study nutritional stress, in contraposition to the consumption of polyfloral pollen. Honeybees feed with Eucalyptus grandis pollen showed a lower abundance of Lactobacillus mellifer and Lactobacillus apis (Firm-4 and Firm-5, respectively) and Bifidobacterium spp. and a higher abundance of Bartonella apis, than honeybees fed with polyfloral pollen. Besides the impact of nutritional stress on honeybee microbiota, it also decreased the expression levels of vitellogenin and genes associated to immunity (glucose oxidase, hymenoptaecin and lysozyme). Finally, Eucalyptus grandis pollen favored the multiplication of Nosema ceranae. These results show that nutritional stress impacts the honeybee gut microbiota, having consequences on honeybee immunity and pathogen development. Those results may be useful to understand the influence of modern agriculture on honeybee health.






Similar content being viewed by others
References
Klein AM, Vaissiere BE, Cane JH, Steffan-Dewenter I, Cunningham SA, Kremen C, Tscharntke T (2007) Importance of pollinators in changing landscapes for world crops. Proc R Soc B Biol Sci 274:303–313. https://doi.org/10.1098/rspb.2006.3721
Potts SG, Imperatriz-Fonseca V, Ngo HT, Aizen MA, Biesmeijer JC, Breeze TD, Dicks LV, Garibaldi LA, Hill R, Settele J, Vanbergen AJ (2016) Safeguarding pollinators and their values to human well-being. Nature 540:220–229. https://doi.org/10.1038/nature20588
Neumann P, Carreck NL (2010) Honey bee colony losses. J Apic Res 49:1–6. https://doi.org/10.3896/IBRA.1.49.1.01
Kulhanek K, Steinhauer N, Rennich K, Caron DM, Sagili RR, Pettis JS, Ellis JD, Wilson ME, Wilkes JT, Tarpy DR, Rose R, Lee K, Rangel J, vanEngelsdorp D (2017) A national survey of managed honey bee 2015-2016 annual colony losses in the USA. J Apic Res 56:328–340. https://doi.org/10.1080/00218839.2017.1344496
Gray A, Brodschneider R, Adjlane N, Ballis A, Brusbardis V, Charrière J-D, Chlebo R, Coffey M F, Cornelissen B, Amaro da Costa C, Csáki T, Dahle B, Danihlík J, Maja Dražić M, Evans G, Fedoriak M, Forsythe I, de Graaf D, Gregorc A, Johannesen J, Kauko L, Kristiansen P, Martikkala M, Martín-Hernández R, Medina-Flores C A, Mutinelli F, Patalano S, Petrov P, Raudmets A, Ryzhikov VA, Simon-Delso N, Stevanovic J, Topolska G, Uzunov A, Vejsnaes F, Williams A, Zammit-Mangion M, Soroker V (2019) Loss rates of honey bee colonies during winter 2017/18 in 36 countries participating in the COLOSS survey, including effects of forage sources, J Apic Res, 58:4, 479–485. https://doi.org/10.1080/00218839.2019.1615661
Requier F, Antúnez K, Morales CL, Aldea Sánchez P, Castilhos D, Garrido M, Giacobino A, Reynaldi FJ, Rosso Londoño JM, Santos E, Garibaldi LA (2018) Trends in beekeeping and honey bee colony losses in Latin America. J Apic Res 57:657–662. https://doi.org/10.1080/00218839.2018.1494919
Goulson D, Nicholls E, Botias C, Rotheray EL (2015) Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347. https://doi.org/10.1126/science.1255957
Steinhauer N, Kulhanek K, Antúnez K, Human H, Chantawannakul P, Chauzat MP, vanEngelsdorp D (2018) Drivers of colony losses. Curr Opin Insect Sci 26:142–148. https://doi.org/10.1016/j.cois.2018.02.004
Naug D (2009) Nutritional stress due to habitat loss may explain recent honeybee colony collapses. Biol Conserv 142:2369–2372. https://doi.org/10.1016/j.biocon.2009.04.007
Keller I, Fluri P, Imdorf A (2005) Pollen nutrition and colony development in honey bees: part I. Bee World 86:3–10. https://doi.org/10.1080/0005772X.2005.11099641
Brodschneider R, Crailsheim K (2010) Nutrition and health in honey bees. Apidologie 41:278–294. https://doi.org/10.1051/apido/2010012
DeGrandi-Hoffman G, Chen Y, Huang E, Huang MH (2010) The effect of diet on protein concentration, hypopharyngeal gland development and virus load in worker honey bees (Apis mellifera L.). J Insect Physiol 56:1184–1191. https://doi.org/10.1016/j.jinsphys.2010.03.017
Alaux C, Dantec C, Parrinello H, Le Conte Y (2011) Nutrigenomics in honey bees: digital gene expression analysis of pollen's nutritive effects on healthy and varroa-parasitized bees. BMC Genomics 12:496. https://doi.org/10.1186/1471-2164-12-496
Di Pasquale G, Salignon M, Le Conte Y, Belzunces LP, Decourtye A, Kretzschmar A, Suchail S, Brunet JL, Alaux C (2013) Influence of pollen nutrition on honey bee health: do pollen quality and diversity matter? PLoS ONE 8:e72016. https://doi.org/10.1371/journal.pone.0072016
Alaux C, Ducloz F, Crauser D, Le Conte Y (2010) Diet effects on honeybee immunocompetence. Biol Lett 6:562–565. https://doi.org/10.1098/rsbl.2009.0986
Schmidt JO, Thoenes SC, Levin MD (1987) Survival of honey bees, Apis mellifera (Hymenoptera: Apidae), fed various pollen sources. J Econ Entomol 80:176–183. https://doi.org/10.1093/aesa/80.2.176
Rinderer TE, Elliott KD (1977) Worker honey bee response to infection with Nosema apis. J Econ Entomol 70:431–433. https://doi.org/10.1093/jee/70.4.431
Porrini MP, Sarlo E, Medici SM, Garrido PM, Porrini DP, Damiani N, Eguaras MJ (2011) Nosema ceranae development in Apis mellifera: influence of diet and infective inoculums. J Apic Res 50:35–41. https://doi.org/10.3896/IBRA.1.50.1.04
Basualdo M, Barragán S, Antúnez K (2014) Bee bread increases honeybee haemolymph protein and promote better survival despite of causing higher Nosema ceranae abundance in honeybees. Environ Microbiol Rep 6:396–400. https://doi.org/10.1111/1758-2229.12169
Branchiccela B, Castelli L, Corona M, Díaz-Cetti S, Invernizzi C, de la Escalera MG, Mendoza Y, Santos E, Silva C, Zunino P, Antúnez K (2019) Impact of nutritional stress on the honeybee colony health. Sci Rep 12:9:10156. https://doi.org/10.1038/s41598-019-46453-9
Invernizzi C, Santos E, García E, Daners G, Di Landro R, Saadoun A, Cabrera C (2011) Sanitary and nutritional characterization of honeybee colonies in Eucalyptus grandis plantations. Arch zootec 60:1303–1314. https://doi.org/10.4321/S0004-05922011000400045
Mendoza Y, Díaz S, Ramallo G, Invernizzi C (2012) Incidence of Nosema ceranae during winter in honey bees colonies removed from Eucaliptus grandis plantations. Veterinaria 48(188):13–19
Roulston TH, Cane JH (2000) Pollen nutritional content and digestibility for animals. Plant Syst Evol 222:187–209. https://doi.org/10.1007/BF00984102
Arien Y, Dag A, Zarchin S, Masci T, Shafira S (2015) Omega-3 deficiency impairs honey bee learning. Proc Natl Acad Sci of the U S A 51:15761–15766. https://doi.org/10.1073/pnas.1517375112
Groot D (1953) Protein and amino acid requirements of the honey bee (Apis mellifera L.). Physiol Comp Oecol 3:197–285
Somerville DC (2001) Nutritional value of bee collected pollens. Rural Industries Research and Development Corporation 1–166
Jones JC, Fruciano C, Hildebrand F, Al Toufalilia H, Balfour NJ, Bork P, Engel P, Ratnieks FLW, Hughes WOH (2018) Gut microbiota composition is associated with environmental landscape in honey bees. Ecol Evol 8:441–451. https://doi.org/10.1002/ece3.3597
Sabree ZL, Hansen AK, Moran NA (2012) Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PLoS One 7:e41250. https://doi.org/10.1371/journal.pone.0041250
Moran NA, Hansen AK, Powell JE, Sabree ZL (2012) Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7:e36393. https://doi.org/10.1371/journal.pone.0036393
Raymann K, Moran NA (2018) The role of the gut microbiome in health and disease of adult honey bee workers. Curr Opin Insect Sci 26:97–104. https://doi.org/10.1016/j.cois.2018.02.012
Engel P, Martinson VG, Moran NA (2013) Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A 109:11002–11007. https://doi.org/10.5281/zenodo.147966
Kwong WK, Moran NA (2013) Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int J Syst Evol Microbiol 63:2008–2018. https://doi.org/10.1099/ijs.0.044875-0
Ellegaard KM, Tamarit D, Javelind E, Olofsson TC, Andersson SG, Vasquez A (2015) Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genomics 16:284. https://doi.org/10.1186/s12864-015-1476-6
Engel P, Kwong WK, Moran NA (2013) Frischella perrara gen. Nov., sp. nov., a gammaproteobacterium isolated from the gut of the honeybee, Apis mellifera. Int J Syst Evol Microbiol 63:3646–3651. https://doi.org/10.1099/ijs.0.049569-0
Kesnerova L, Moritz R, Engel P (2016) Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int J Syst Evol Microbiol 66:414–421. https://doi.org/10.1099/ijsem.0.000736
Corby-Harris V, Snyder LA, Schwan MR, Maes P, McFrederick QS, Anderson KE (2014a) Origin and effect of alpha 2.2 Acetobacteraceae in honey bee larvae and description of Parasaccharibacter apium gen. Nov., sp. nov. Appl Environ Microbiol 80:7460–7472. https://doi.org/10.1128/AEM.02043-14
Evans JD, Spivak M (2010) Socialized medicine: individual and communal disease barriers in honey bees. J Invertebr Pathol 103:S62–S72. https://doi.org/10.1016/j.jip.2009.06.019
Danihlík J, Aronstein K, Petřivalský M (2016) Antimicrobial peptides: a key component of honey bee innate immunity. J Apic Res 54:123–136. https://doi.org/10.1080/00218839.2015
White JWJ, Subers MH, Schepartz AI (1963) The identification of inhibine, antibacterial factor in honey, as hydrogen peroxide, and its origin in a honey glucose oxidase system. Biochim Biophys Acta 73:57–70. https://doi.org/10.1016/0926-6569(63)90108-1
Doublet V, Poeschl Y, Gogol-Döring A, Alaux C, Annoscia D, Aurori C, Barribeau SM, Bedoya-Reina OC, Brown MJ, Bull JC, Flenniken ML, Galbraith DA, Genersch E, Gisder S, Grosse I, Holt HL, Hultmark D, Lattorff HM, Le Conte Y, Manfredini F, McMahon DP, Moritz RF, Nazzi F, Niño EL, Nowick K, van Rij RP, Paxton RJ, Grozinger CM (2017) Unity in defense: honeybee workers exhibit conserved molecular responses to diverse pathogens. BMC Genomics 18:207. https://doi.org/10.1186/s12864-017-3597-6
Faegri K, Iversen J (1975) Textbook of modern pollen analysis. T Munksgaard Copenhagen, p 423
Louveaux J, Mauricio A, Vorwohl G (1978) Methods of melissopalynology. Bee World 59:139–157. https://doi.org/10.1080/0005772X.1978.11097714
Wiliams GR, Alaux C, Costa C, Csákit T, Doublet V, Eisenhardt D, Fries I, Kuhn R, Mcmahon DP, Medrzycki P, Murray TE, Natsopoulou ME, Neumann, P, Oliver R, Paxton RJ, Pernal SF, Shutler D, Tanner G, Van der steen JJM, Brodschneider R (2013) Standard methods for maintaining adult Apis mellifera in cages under in vitro laboratory conditions. In V Dietemann, JD Ellis, P Neumann (Eds) the COLOSS BEEBOOK, volume I: standard methods for Apis mellifera research. J Apic Res 52(1). https://doi.org/10.3896/IBRA.1.52.1.04
Porrini MP, Garrido PM, Eguaras MJ (2013) Individual feeding of honeybees:modification of the Rinderer technique. J Apic Res 52:194–195
Fries I, Chauzat MP, Chen YP, Doublet V, Genersch E, Gisder S, Higes M, McMahon DP, Martín-Hernández R, Natsopoulou M, Paxton RJ, Tanner G, Webster TC, Williams GR (2013) Standard methods for Nosema research. J Apic Res 52:1–28. https://doi.org/10.3896/IBRA.1.52.1.14
Human H, Beodschneider R, Dietemann V, Dively G, Ellis J, Forsgren E, Fries I, Hatjina F, Hu F-L, Jaffé R, Köhler A, Pirk CWW, Rose R, Strauss U, Tanner G, Van der Steen JJM, Vejsnaes F, Williams GR, Zheng H-Q (2013) Miscellaneous standard methods for Apis mellifera research. In V Dietemann; J D Ellis; P Neumann (Eds) the COLOSS BEEBOOK, volume I: standard methods for Apis mellifera research. J Apic Res 52(4). https://doi.org/10.3896/IBRA.1.52.4.10
Martin-Hernandez R, Meana A, Prieto L, Salvador AM, Garrido-Bailon E, Higes M (2007) Outcome of colonization of Apis mellifera by Nosema ceranae. Appl Environ Microbiol 73:6331–6338. https://doi.org/10.1128/AEM.00270-07
Anderson KE, Carroll MJ, Sheehan T, Lanan MC, Mott BM, Maes P, Corby-Harris V (2014) Hive-stored pollen of honey bees: many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol Ecol 23:5904–5917. https://doi.org/10.1111/mec.12966
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322 0099-2240/96/$04.0010
Arismendi N, Bruna A, Zapata N, Vargas M (2016) PCR-specific detection of recently described Lotmaria passim (Trypanosomatidae) in Chilean apiaries. J Invertebr Pathol 134:1–5. https://doi.org/10.1016/j.jip.2015.12.008
Denman SE, McSweeney CS (2006) Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol 58:572–582. https://doi.org/10.1111/j.1574-6941.2006.00190.x
Evans JD (2006) Beepath: an ordered quantitative-PCR array for exploring honey bee immunity and disease. J Invertebr Pathol 93:135–139. https://doi.org/10.1016/j.jip.2006.04.004
Ott SJ, Musfeldt M, Ullmann U, Hampe J, Schreiber S (2004) Quantification of intestinal bacterial populations by real-time PCR with a universal primer set and minor groove binder probes: a global approach to the enteric Flora. J Clin Microbiol 42:2566–2572. https://doi.org/10.1128/JCM.42.6.2566-2572.2004
Yang X, Cox-Foster DL (2005) Impact of an ectoparasite on the immunity and pathology of an invertebrate: evidence for host immunosuppression and viral amplification. Proc Natl Acad Sci U S A 102:7470–7475. https://doi.org/10.1073/pnas.0501860102
Corona M, Velarde R, Remolina S, Moran-Lauter A, Wang Y, Hughes KA, Robinson GE (2007) Vitellogenin, juvenile hormone, insulin signalling, and queen honey bee longevity. Proc Natl Acad Sci U S A 104:7128–7133. https://doi.org/10.1073/pnas.0701909104
Engel P, James RR, Koga R, Kwong WK, McFrederick QS, Moran NA (2013) Standard methods for research on Apis mellifera gut symbionts. J Apic Res 52(4):1–24. https://doi.org/10.3896/IBRA.1.52.4.07
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Lan Y, Wang Q, Cole JR, Rosen GL (2012) Using the RDP classifier to predict taxonomic novelty and reduce the search space for finding novel organisms. Public Library of Science ONE 7:e32491. https://doi.org/10.1371/journal.pone.0032491
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
R Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(e45):387–409. https://doi.org/10.1051/apido:2000130
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron:4 http://palaeo-electronica.org/2001_2001/past/issue2001_2001.htm
Powell JE, Martinson VG, Urban-Mead K, Moran NA (2014) Routes of Acquisition of the gut microbiota of the honey bee Apis mellifera. Appl Environ Microbiol 80:7378–7387. https://doi.org/10.1128/AEM.01861-14
Bottacini F, Milani C, Turroni F, Sánchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, Delledonne M, Henrissat B, Coutinho P, Fitzgerald GF, Margolles A, van Sinderen VM (2012) Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7(9):e44229. https://doi.org/10.1371/journal.pone.0044229
Kwong WK, Moran NA (2016) Gut microbial communities of social bees. Nat Rev Microbiol 14:374–384. https://doi.org/10.1038/nrmicro.2016.43
Forsgren E, Olofsson TC, Vasquez A, Fries I (2010) Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 41:99–108. https://doi.org/10.1051/apido/2009065
Vásquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, Olofsson TC (2012) Symbionts as major modulators of insect health: lactic acid Bacteria and honeybees. PLoS One 7(7):e33188. https://doi.org/10.1371/journal.pone.0033188
Killer J, Dubná S, Sedláček I, Švec P (2014) Lactobacillus apis sp. nov., from the stomach of honeybees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. Int J Syst Evol Microbiol 64:152–157. https://doi.org/10.1099/ijs.0.053033-0
Chomel BB, Kasten RW (2010) Bartonellosis, an increasingly recognized zoonosis. J Appl Microbiol 109:743–750. https://doi.org/10.1111/j.1365-2672.2010.04679.x
Kopecky J, Nesvorna M, Hubert J (2014) Bartonella-like bacteria carried by domestic mite species. Exp Appl Acarol 64:21–32. https://doi.org/10.1007/s10493-014-9811-1
Hubert J, Erban T, Kamler M, Kopecky J, Nesvorna M, Hejdankova S, Titera D, Tyl J, Zurek L (2015) Bacteria detected in the honeybee parasitic mite Varroa destructor collected from beehive winter debris. J Appl Microbiol 119:640–654. https://doi.org/10.1111/jam.12899
Amdam GV, Norberg K, Hagen A, Omholt SW (2003) Social exploitation of vitellogenin. Proc Natl Acad Sci USA 100: 1799-1802. https://doi.org/10.1073/pnas.0333979100
Seehuus SC, Norberg K, Gimsa U, Krekling T, Amdam GV (2006) Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proceedings of the National Academy of Sciences USA 103:962–967. https://doi.org/10.1073/pnas.0502681103
Nelson CM, Ihle KE, Fondrk MK, Page Jr RE, Amdam GV (2007) The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biology 5: 0673–0677. https://doi.org/10.1371/journal.pbio.0050062
Bitondi MMG, Simões ZLP (1996) The relationship between level of pollen in the diet, vitellogenin and juvenile hormone titres in Africanized Apis mellifera workers. J Apic Res 35:27–36. https://doi.org/10.1080/00218839.1996.11100910
Basualdo M, Barragán S, Vanagas L, García C, Solana H, Rodríguez E, Bedascarrasbure E (2013) Conversion of high and low pollen protein diets into protein in worker honey bees (Hymenoptera: Apidae). J Econ Entomol 106:1553–1558. https://doi.org/10.1603/EC12466
Corby-Harris V, Jones BM, Walton A, Schwan MR, Anderson KE (2014) Transcriptional markers of sub-optimal nutrition in developing Apis mellifera nurse workers. BMC Genomics 15:134. https://doi.org/10.1186/1471-2164-15-134
Casteels P, Ampe C, Jacobs F, Tempst P (1993) Functional and chemical characterization of Hymenoptaecin, an antibacterial polypeptide that is infection-inducible in the honeybee (Apis mellifera). J Biol Chem 268:7044–7054
Gillespie JP, Kanost M (1997) Biological mediators of insect immunity. Annu Rev Entomol 42:611–643. https://doi.org/10.1146/annurev.ento.42.1.611
Imler JL, Bulet P (2005) Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem Immunol Allergy 86:1–21. https://doi.org/10.1159/000086648
Maggi M, Negri P, Plischuk S, Szawarski N, DePiano F, De Feudis L, Eguaras M, Audisio C (2013) Effects of the organic acids produced by a lactic acid bacterium in Apis mellifera colony development, Nosema ceranae control and fumagillin efficiency. Vet Microbiol https://doi.org/10.1016/j.vetmic.2013.07.030
Baffoni L, Gaggìa F, Alberoni D, Cabbri R, Nanetti A, Biavati B, Di Gioia D (2015) Effect of dietary supplementation of Bifidobacterium and Lactobacillus strains in Apis mellifera L. against Nosema ceranae. Benef Microbes 7:1–8. https://doi.org/10.3920/BM2015.0085
Arredondo D, Castelli L, Porrini M, Garrido M, Eguaras M, Zunino P, Antúnez K (2017) Lactobacillus kunkeei strains decreased the infection by honey bee pathogens Paenibacillus larvae and Nosema ceranae. Benef Microbes 9:1–12. https://doi.org/10.3920/BM2017.0075
Rubanov A, Russell KA, Rothman JA, Nieh JC, McFrederick QS (2019) Intensity of Nosema ceranae infection is associated with specific honey bee gut bacteria and weakly associated with gut microbiome structure. Sci Rep 9:3820. https://doi.org/10.1038/s41598-019-40347-6
Antúnez K, Martín-Hernández R, Prieto L, Meana A, Zunino P, Higes M (2009) Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ Microbiol 11:2284–2290. https://doi.org/10.1111/j.1462-2920.2009.01953.x
Mayack C, Naug D (2009) Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection. J Invertebr Pathol 100:185–188. https://doi.org/10.1016/j.jip.2008.12.001
Acknowledgments
This work was supported by “Comisión Sectorial de Investigación Científica” (CSIC 6406) and “Agencia Nacional de Investigación e Innovación” (POS_NAC_2013_1_12228), Uruguay. Authors thank researchers and technicians from the Instituto Nacional de Investigación Agropecuaria (Yamandú Mendoza, Gustavo Ramallo, Carlos Silva and Sebastián Díaz) for their help with sample collection. Finally, we thank the anonymous reviewers for their valuable comments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(PDF 631 kb)
Rights and permissions
About this article
Cite this article
Castelli, L., Branchiccela, B., Garrido, M. et al. Impact of Nutritional Stress on Honeybee Gut Microbiota, Immunity, and Nosema ceranae Infection. Microb Ecol 80, 908–919 (2020). https://doi.org/10.1007/s00248-020-01538-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01538-1