Abstract
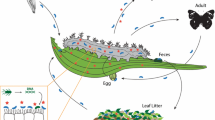
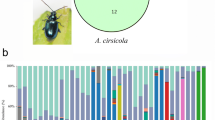
The adaptability of herbivorous insects to toxic plant defense compounds is partly related to the structure of the gut microbiome. To overcome plant resistance, the insect gut microbiome should respond to a wide range of allelochemicals derived from dietary niches. Nevertheless, for sibling herbivorous insect species, whether the gut microbiome contributes to success in food niche competition is unclear. Based on 16S rDNA high-throughput sequencing, the gut microbiomes of two Apriona species that share the same food niche were investigated in this study to determine whether the gut microbiome contributes to insect success in food-niche competition. Our observations indicated that the gut microbiome tended to play a part in host niche competition between the two Apriona species. The gut microbiome of Apriona swainsoni had many enriched pathways that can help degrade plant toxic secondary compounds, including xenobiotic biodegradation and metabolism, terpenoid and polyketide metabolism, and secondary metabolite biosynthesis. Meanwhile, A. swainsoni hosted a much greater variety of microorganisms and had more viable bacteria than A. germari. We conclude that gut microbes may influence the coevolution of herbivores and host plants. Gut bacteria may not only serve to boost nutritional relationships, but may also play an important role in insect food niche competition.






Similar content being viewed by others
References
Futuyma DJ, Moreno G (1988) The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 19:207–233
Bird G, Kaczvinsky C, Wilson AE, Hardy NB (2019) When do herbivorous insects compete? A phylogenetic meta-analysis. Ecol. Lett. 22:875–883
Resetarits WJ, Pintar MR, Bohenek JR, Breech TM (2019) Patch size as a niche dimension: aquatic insects behaviorally partition enemy-free space across gradients of patch size. Am. Nat. 194:776–793
Mirabasso J, Bissattini AM, Bologna MA, Luiselli L, Stellati L, Vignoli L (2020) Feeding strategies of co-occurring newt species across different conditions of syntopy: a test of the “within-population niche variation” hypothesis. Diversity 12:181
Theunissen J (1994) Intercropping in field vegetable crops: pest management by agrosystem diversification—an overview. J. Pestic. Sci. 42:65–68
Shaw B, Brain P, Wijnen H, Fountain MT (2018) Reducing Drosophila suzukii emergence through inter-species competition. Pest Manag. Sci. 74:1466–1471
Brévault T, Clouvel P (2019) Pest management: reconciling farming practices and natural regulations. Crop Prot. 115:1–6
Falahzadah MH, Karimi J, Gaugler R (2020) Biological control chance and limitation within integrated pest management program in Afghanistan. Egyp J Biol Pest Co 30:1–10
Awmack CS, Leather SR (2002) Host plant quality and fecundity in herbivorous insects. Annu. Rev. Entomol. 47:817–844
Hardy NB, Kaczvinsky C, Bird G, Normark BB (2020) What we don't know about diet-breadth evolution in herbivorous insects. Annu Rev Ecol Evol Syst. https://doi.org/10.1146/annurev-ecolsys-011720-023322
Toju H, Abe H, Ueno S, Miyazawa Y, Taniguchi F, Sota T, Yahara T (2011) Climatic gradients of arms race coevolution. Am. Nat. 177:562–573
Mopper S (1996) Adaptive genetic structure in phytophagous insect populations. Trends Ecol. Evol. 11:235–238
Zhen Y, Aardema ML, Medina EM, Schumer M, Andolfatto P (2012) Parallel molecular evolution in an herbivore community. Science 337:1634–1637
Dobler S, Petschenka G, Wagschal V, Flacht L (2015) Convergent adaptive evolution–how insects master the challenge of cardiac glycoside-containing host plants. Entomol Exp Appl 157:30–39
Okamura Y, Sato A, Tsuzuki N, Murakami M, Heidel-Fischer H, Vogel H (2019) Molecular signatures of selection associated with host plant differences in Pieris butterflies. Mol. Ecol. 28:4958–4970
Giordano R, Donthu RK, Zimin A, Chavez ICJ, Gabaldon T, van Munster M et al (2020) Soybean aphid biotype 1 genome: insights into the invasive biology and adaptive evolution of a major agricultural pest. Insect Biochem Mol Biol. https://doi.org/10.1016/j.ibmb.2020.103334
Hammer TJ, Bowers MD (2015) Gut microbes may facilitate insect herbivory of chemically defended plants. Oecologia 179:1–14
Shaw BD, Christeller JT (2009) Characterization of the proteases in the midgut of the xylophagous larvae of Oemona hirta (Coleoptera: Cerambycidae). Insect Sci 16:381–386
Faccoli M, Favaro R (2016) Host preference and host colonization of the Asian longhorn beetles, Anoplophora glabripennis (Coleoptera Cerambycidae), in southern Europe. B Entomol Res 106:359
Wang L, Qu L, Zhang L, Hu J, Tang F, Lu M (2016) Metabolic responses of poplar to Apripona germari (Hope) as revealed by metabolite profiling. Int. J. Mol. Sci. 17:923
Li S (2016) Damage of Apriona swainsoni (Hope) on Sophora japonica Linn, in Pingdingshan area. Plant Dis Pests 7:16–18
Rim K, Golec JR, Duan JJ (2018) Host selection and potential non-target risk of Dastarcus helophoroides, a larval parasitoid of the Asian longhorned beetle, Anoplophora glabripennis. Biol. Control 123:120–126
Ma Y, Shi L, Zhao Y, Xu H (2018) Comparison of volatiles released from the host Juglans mandshurica in different damaged states and the GC-EAD and behavioral responses of Apriona germari (Coleoptera: Cerambycidae) to these volatiles. Acta Entom Sinica 61:574–584
Cai Y, Qiu F, Huang K, Zhang X, Hong W, Huang J (2020) Microbiological safety evaluation of edible bucket worms. J Agri Biot 28:1675–1687
Zhang SK, Shu JP, Xue HJ, Zhang W, Zhang YB, Liu YN, Fang LX, Wang YD, Wang HJ (2020) The gut microbiota in camellia weevils are influenced by plant secondary metabolites and contribute to saponin degradation. mSystems 5. https://doi.org/10.1128/mSystems.00692-19
Rattan RS (2010) Mechanism of action of insecticidal secondary metabolites of plant origin. Crop Prot. 29:913–920
Sengottayan SN (2013) Physiological and biochemical effect of neem and other Meliaceae plants secondary metabolites against lepidopteran insects. Front. Physiol. 4:359
AlJabr AM, Hussain A, Rizwan-ul-Haq M, Al-Ayedh H (2017) Toxicity of plant secondary metabolites modulating detoxification genes expression for natural red palm weevil pesticide development. Molecules 22:169
Stevenson P (2020) For antagonists and mutualists: the paradox of insect toxic secondary metabolites in nectar and pollen. Phytochem. Rev. 19:603–614
Yactayo-Chang JP, Tang HV, Mendoza J, Christensen SA, Block AK (2020) Plant defense chemicals against insect pests. Agronomy 10:1156
Carmona D, Lajeunesse MJ, Johnson MT (2011) Plant traits that predict resistance to herbivores. Funct. Ecol. 25:358–367
Mithöfer A, Boland W (2012) Plant defense against herbivores: chemical aspects. Annu. Rev. Plant Biol. 63:431–450
Turnbull LA, Isbell F, Purves DW, Loreau M, Hector A (2016) Understanding the value of plant diversity for ecosystem functioning through niche theory. P Roy Soc B: Biol Sci 283:20160536
Amato KR, Sanders JG, Song SJ, Nute M, Metcalf JL, Thompson LR, Morton JT, Amir A, McKenzie VJ, Humphrey G, Gogul G, Gaffney J, Baden AL, Britton GAO, Cuozzo FP, Di Fiore A, Dominy NJ, Goldberg TL, Gomez A, Kowalewski MM, Lewis RJ, Link A, Sauther ML, Tecot S, White BA, Nelson KE, Stumpf RM, Knight R, Leigh SR (2019) Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiome. ISME J 13:576–587
Gupta A, Nair S (2020) Dynamics of insect-microbiome interaction influence host and microbial symbiont. Front. Microbiol. 11:1357
Douglas AE (2015) Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60:17
Kundu P, Manna B, Majumder S, Ghosh A (2019) Species-wide metabolic interaction network for understanding natural lignocellulose digestion in termite gut microbiota. Sci. Rep. 9:1–13
Zhang ZJ, Huang MF, Qiu LF, Song RH, Zhang ZX, Ding YW, Zhou X, Zhang X, Zheng H (2020) Diversity and functional analysis of Chinese bumblebee gut microbiota reveal the metabolic niche and antibiotic resistance variation of Gilliamella. Insect Sci. https://doi.org/10.1111/1744-7917.12770
Wink M (2018) Plant secondary metabolites modulate insect behavior-steps toward addiction? Front. Physiol. 9:364
Alberdi A, Aizpurua O, Bohmann K, Zepeda-Mendoza ML, Gilbert MTP (2016) Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31:689–699
Chen S, Zhou Y, Chen Y, Gu J (2018) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890
Zhang SK, Shu JP, Xue HJ, Zhang W, Wang HJ (2018) Genetic diversity in the camellia weevil, Curculio chinensis Chevrolat (Coleoptera: Curculionidae) and inferences for the impact of host plant and human activity. Entomol Sci 21:447–460
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Dixon P (2003) VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196
Domik D, Thürmer A, Weise T, Brandt W, Daniel R, Piechulla B (2016) A terpene synthase is involved in the synthesis of the volatile organic compound sodorifen of Serratia plymuthica 4Rx13. Front. Microbiol. 7:737
Wickham H (2011) ggplot2. Wiley Interdisciplinary Reviews: Computational Statistics 3:180–185
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645
Kolde R, Kolde MR (2015) Package ‘pheatmap’. R Package 1:790
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol. 12:1–18
Csardi MG (2013) Package ‘igraph’. Last accessed 3:2013
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10:57–59
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267
Robin F, Dubois C, Curti D, Schuchmann HP, Palzer S (2011) Effect of wheat bran on the mechanical properties of extruded starchy foams. Food Res. Int. 44:2880–2888
Hamilton NE, Ferry M (2018) Ggtern: ternary diagrams using ggplot2. J. Stat. Softw. 87:1–17
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16s rDNA marker gene sequences. Nat. Biotechnol. 31:814–821
Ward T, Larson J, Meulemans J, Hillmann B, Lynch J, Sidiropoulos D, Spear JR, Caporaso G, Blekhman R, Knight R, Fink R, Knights D (2017) BugBase predicts organism-level microbiome phenotypes. BioRxiv p.133462
Bekir J, Mars M, Vicendo P, Fterrich A, Bouajila J (2013) Chemical composition and antioxidant, anti-inflammatory, and antiproliferation activities of pomegranate (Punica granatum) flowers. J. Med. Food 2013(16):544–550
Sheng Z, Wan P, Dong C, Li Y (2013) Optimization of total flavonoids content extracted from Flos Populi using response surface methodology. Ind. Crop. Prod. 43:778–786
Tomar O (2019) The effects of probiotic cultures on the organic acid content, texture profile and sensory attributes of Tulum cheese. Int. J. Dairy Technol. 72:218–228
Wang Z, Wen J, Xing J, He Y (2006) Quantitative determination of diterpenoid alkaloids in four species of Aconitum by HPLC. J Pharmaceut Biomed 40:1031–1034
Itoh H, Tago K, Hayatsu M, Kikuchi Y (2018) Detoxifying symbiosis: microbe-mediated detoxification of phytotoxins and pesticides in insects. Nat. Prod. Rep. 35:434–454
Friberg M, Posledovich D, Wiklund C (2015) Decoupling of female host plant preference and offspring performance in relative specialist and generalist butterflies. Oecologia 178:1181–1192
Kelly CA, Bowers MD (2016) Preference and performance of generalist and specialist herbivores on chemically defended host plants. Ecol Entom 41:308–316
Morera-Margarit P, Pope TW, Mitchell C, Karley AJ (2020) Could bacterial associations determine the success of weevil species? Ann Appl Biol doi. https://doi.org/10.1111/aab.12625
Vanbergen AJ, Espíndola A, Aizen MA (2018) Risks to pollinators and pollination from invasive alien species. Nat Ecol Evol 2:16–25
Kostovcik M, Bateman CC, Kolarik M, Stelinski LL, Jordal BH, Hulcr J (2015) The ambrosia symbiosis is specific in some species and promiscuous in others: evidence from community pyrosequencing. ISME J 9:126–138
Cheng C, Wickham JD, Chen L, Xu D, Lu M, Sun J (2018) Bacterial microbiota protect an invasive bark beetle from a pine defensive compound. Microbiome 6:132
Mason CJ, Jones AG, Felton GW (2019) Co-option of microbial associates by insects and their impact on plant–folivore interactions. Plant Cell Environ. 42:1078–1086
David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563
Morales-Jiménez J, Zúñiga G, Villa-Tanaca L, Hernández-Rodríguez C (2009) Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae). Microb. Ecol. 58:879–891
Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230
Hansen AK, Moran NA (2014) The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 23:1473–1496
Mason CJ, Ray S, Shikano I, Peiffer M, Jones AG, Luthe DS, Hoover K, Felton GW (2019) Plant defenses interact with insect enteric bacteria by initiating a leaky gut syndrome. P Natl A Sci 116:15991–15996
Wei J, Segraves KA, Li WZ, Yang XK, Xue HJ (2020) Gut bacterial communities and their contribution to performance of specialist Altica flea beetles. Microb. Ecol. 80:946–959
Acknowledgements
We thank Wang Y. for helping with longhorn beetle collection, Prof. J. P. S. for valuable discussions. S. K. Z., Y. W. contributed fieldwork. S. K. Z., Y. W. and Z. K. L. performed the laboratory experiments and analyzed the data. X. D. Z. and J. H. H. coordinated the study and participated in conceptual design and manuscript preparation. S. K. Z., W.Y. performed most of the work for conceptual design and manuscript preparation. All authors read and approved the final manuscript.
Funding
This work was supported by the Cooperation Project of Zhejiang Province and Chinese Academy of Forestry (Grant No. 2020SY08).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Zhang, SK., Wang, Y., Li, ZK. et al. Two Apriona Species Sharing a Host Niche Have Different Gut Microbiome Diversity. Microb Ecol 83, 1059–1072 (2022). https://doi.org/10.1007/s00248-021-01799-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01799-4