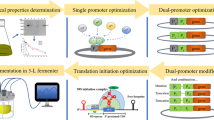
Abstract
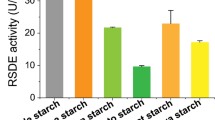
Raw starch-degrading enzymes (RSDEs) are capable of directly degrading raw starch granules below the gelatinization temperature of starch, which may significantly reduce the cost of starch-based biorefining. However, low yields of natural RSDEs from filamentous fungi limit their industrial application. In this study, transcriptomic and secretomic profiling was employed to screen strongest promoters and signal peptides for use in overexpression of a RSDE gene in Penicillium oxalicum. Top five strong promoters and three signal peptides were detected. Using a green fluorescent protein (GFP) as the reporter, the inducible promoter pPoxEgCel5B of an endoglucanase gene PoxEgCel5B and the signal peptide spPoxGA15A of a raw starch-degrading glucoamylase PoxGA15A were respectively identified as driving the highest GFP production in P. oxalicum. PoxGA15A-overexpressed P. oxalicum strain OXPoxGA15A, which was constructed based on both pPoxEgCel5B and spPoxGA15A, produced significantly higher amounts of recombinant PoxGA15A than the parental strain ∆PoxKu70. Furthermore, crude enzyme from the OXPoxGA15A strain exhibited high activities towards raw starch from cassava, potato, and uncooked soluble starch. Specifically, raw cassava starch-degrading enzyme activity reached 241.6 U/mL in the OXPoxGA15A, which was 3.4-fold higher than that of the ∆PoxKu70. This work provides a feasible method for hyperproduction of RSDEs in P. oxalicum.




Similar content being viewed by others
References
Ayodeji AO, Ogundolie FA, Bamidele OS, Kolawole AO, Ajele JO (2017) Raw starch degrading, acidic-thermostable glucoamylase from Aspergillus fumigatus CFU-01: purification and characterization for biotechnological application. J Microbiol Biotechnol 6:90–100
Božić NLN, Slavić MŠ, Vujčić Z (2017) Raw starch degrading α-amylases: an unsolved riddle. Amylase 1:12–25
Garvey M, Klose H, Fischer R, Lambertz C, Commandeur U (2013) Cellulases for biomass degradation: comparing recombinant cellulase expression platforms. Trends Biotechnol 31:581–593
Gomes E, Souza SRD, Grandi RP, Silva RD (2005) Production of thermostable glucoamylase by newly isolated Aspergillus flavus A 1.1 and Thermomyces lanuginosus A 13.37. Braz J Microbiol 36:75–82
Haasum I, Eriksen SH, Jensen B (1991) Growth and glucoamylase production by the thermophilic fungus Thermophilus lanuginose in a synthetic medium. Appl Microbiol Biotechnol 34(5):656–660
Hu YB, Xue HZ, Liu GD, Song X, Qu YB (2015) Efficient production and evaluation of lignocellulolytic enzymes using a constitutive protein expression system in Penicillium oxalicum. J Ind Microbiol Biotechnol 42:877–887
Krumm A, Hickey LB, Groudine M (1995) Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev 9:559–572
Kumar S, Satyanarayana T (2001) Medium optimization for glucoamylase production by a yeast, Pichia subpelliculosa ABWF-64, in submerged cultivation. World J Microbiol Biotechnol 17(1):83–87
Kumar S, Kumar P, Satyanarayana T (2007) Production of raw starch-saccharifying thermostable and neutral glucoamylase by the thermophilic mold Thermomucor indicae-seudaticae, in submerged fermentation. Appl Biochem Biotechnol 142(3):221–230
Li H, Chi Z, Duan X, Wang L, Sheng J, Wu L (2007) Glucoamylase production by the marine yeast Aureobasidium pullulans N13d and hydrolysis of potato starch granules by the enzyme. Process Biochem 42(3):462–465
Liao Y, Huang L, Wang B, Zhou F, Pan L (2015) The global transcriptional landscape of Bacillus amyloliquefaciens XH7 and high-throughput screening of strong promoters based on RNA-seq data. Gene 571:252–262
Lin HJ, Xian L, Zhang QJ, Luo XM, Xu QS, Yang Q, Duan CJ, Liu JL, Tang JL, Feng JX (2011) Production of raw cassava starch-degrading enzyme by Penicillium and its use in conversion of raw cassava flour to ethanol. J Ind Microbiol Biotechnol 38:733–742
Liu GD, Zhang L, Qin YQ, Zou G, Li ZH, Yan X, Wei XM, Chen M, Chen L, Zheng K, Zhang J, Ma L, Li J, Liu R, Xu H, Bao XM, Fang X, Wang LS, Zhong YH, Liu WF, Zheng HJ, Wang SY, Wang CS, Xun LY, Zhao GP, Wang TH, Zhou ZH, Qu YB (2013) Long-term strain improvements accumulate mutations in regulatory elements responsible for hyper-production of cellulolytic enzymes. Sci Rep 3:1569
Lomthong T, Chotineeranat S, Kitpreechavanich V (2015) Production and characterization of raw starch degrading enzyme from a newly isolated thermophilic filamentous bacterium, Laceyella sacchari LP175. Starch-Stärke 67:255–266
Lomthong T, Lertwattanasakul N, Kitpreechavanich V (2016) Production of raw starch degrading enzyme by the thermophilic filamentous bacterium Laceyella sacchari LP175 and its application for ethanol production from dried cassava chips. Starch-Stärke 68:1264–1274
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Morita H, Fujio Y (1997) High specific activity of raw-starch digesting-glucoamylase producing Rhizopus, sp. A-11 in liquid culture. Starch-Stärke 49:293–296
Moshi AP, Hosea KMM, Elisante E, Mamo G, Onnby L, Nges IA (2016) Production of raw starch-degrading enzyme by Aspergillus sp and its use in conversion of inedible wild cassava flour to bioethanol. J Biosci Bioeng 121:457–463
Nevalainen H, Peterson R (2014) Making recombinant proteins in filamentous fungi- are we expecting too much? Front Microbiol 5:75
Park J, Hulsman M, Arentshorst M, Breeman M, Alazi E, Lagendijk EL, Rocha MC, Malavazi I, Nitsche BM, van den Hondel CA (2016) Transcriptomic and molecular genetic analysis of the cell wall salvage response of Aspergillus niger to the absence of galactofuranose synthesis. Cell Microbiol 18:1268–1284
Sevillano L, Vijgenboom E, van Wezel GP, Diaz M, Santamaria RI (2016) New approaches to achieve high level enzyme production in Streptomyces lividans. Microb Cell Factories 15:28
Singh A, Taylor LE, Vander Wall TA, Linger J, Himmel ME, Podkaminer K, Adney WS, Decker SR (2015) Heterologous protein expression in Hypocrea jecorina: a historical perspective and new developments. Biotechnol Adv 33:142–154
Sun HY, Zhao PJ, Ge XY, Xia YJ, Hao ZK, Liu JW, Peng M (2010) Recent advances in microbial raw starch degrading enzymes. Appl Biochem Biotechnol 160:988–1003
Xu LL, Shen Y, Hou J, Peng BY, Tang HT, Bao XM (2014) Secretory pathway engineering enhances secretion of cellobiohydrolase I from Trichoderma reesei in Saccharomyces cerevisiae. J Biosci Bioeng 117:45–52
Xu QS, Yan YS, Feng JX (2016) Efficient hydrolysis of raw starch and ethanol fermentation: a novel raw starch-digesting glucoamylase from Penicillium oxalicum. Biotechnol Biofuels 9:216
Xu Q, Knoshaug EP, Wang W, Alahuhta M, Baker JO, Yang SH, Vander WTA, Decker SR, Himmel ME, Zhang M, Wei H (2017) Expression and secretion of fungal endoglucanase II and chimeric cellobiohydrolase I in the oleaginous yeast Lipomyces starkeyi. Microb Cell Factories 16:126
Yan YS, Zhao S, Liao LS, He QP, Xiong YR, Wang L, Li CX, Feng JX (2017) Transcriptomic profiling and genetic analyses reveal novel key regulators of cellulase and xylanase gene expression in Penicillium oxalicum. Biotechnol Biofuels 10:279
Zhang Z, Liu JL, Lan JY, Duan CJ, Ma QS, Feng JX (2014) Predominance of Trichoderma and Penicillium in cellulolytic aerobic filamentous fungi from subtropical and tropical forests in China, and their use in finding highly efficient β-glucosidase. Biotechnol Biofuels 7:107
Zhao S, Yan YS, He QP, Yang L, Yin X, Li CX, Mao LC, Liao LS, Huang JQ, Xie SB, Nong QD, Zhang Z, Jing L, Xiong YR, Duan CJ, Liu JL, Feng JX (2016) Comparative genomic, transcriptomic and secretomic profiling of Penicillium oxalicum HP7-1 and its cellulase and xylanase hyper-producing mutant EU2106, and identification of two novel regulatory genes of cellulase and xylanase gene expression. Biotechnol Biofuels 9:203
Ziegler CM, Eisenhauer P, Bruce EA, Weir ME, King BR, Klaus JP, Krementsov DN, Shirley DJ, Ballif BA, Botten J (2016) The Lymphocytic choriomeningitis virus matrix protein PPXY late domain drives the production of defective interfering particles. PLoS Pathog 12:e1005501
Zou G, Shi SH, Jiang YP, van den Brink J, de Vries RP, Chen L, Zhang J, Ma L, Wang CS, Zhou ZH (2012) Construction of a cellulase hyper-expression system in Trichoderma reesei by promoter and enzyme engineering. Microb Cell Factories 11:12
Funding
This work was supported by the Guangxi BaGui Scholars Program Foundation (Grant No. 2011A001) and the Guangxi Natural Science Foundation (Grant No. 2012GXNSFGA060005) to JXF and the “One Hundred Person” Project of Guangxi to SZ.
Author information
Authors and Affiliations
Contributions
JXF designed and supervised the research and was involved in the data analysis and the preparation of the manuscript. SZ cosupervised all the research and was involved in the analysis of all experimental data and the preparation of the manuscript. LW did the experiments of construction of deletion mutants, overexpressed strains, green fluorescence intensity measurement, and enzyme activity tests and was involved in the preparation of the manuscript. XXC was involved in the screening of candidate promoters and signal peptides in P. oxalicum. QPD was involved in data analysis. CXL performed bioinformatic analysis and primer design. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 1.37 mb)
Rights and permissions
About this article
Cite this article
Wang, L., Zhao, S., Chen, XX. et al. Secretory overproduction of a raw starch-degrading glucoamylase in Penicillium oxalicum using strong promoter and signal peptide. Appl Microbiol Biotechnol 102, 9291–9301 (2018). https://doi.org/10.1007/s00253-018-9307-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9307-8