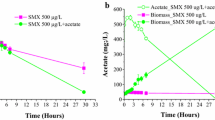
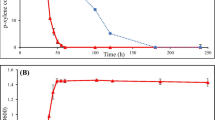
Abstract
In the last decade, biological degradation and mineralization of antibiotics have been increasingly reported feats of environmental bacteria. The most extensively described example is that of sulfonamides that can be degraded by several members of Actinobacteria and Proteobacteria. Previously, we reported sulfamethoxazole (SMX) degradation and partial mineralization by Achromobacter denitrificans strain PR1, isolated from activated sludge. However, further studies revealed an apparent instability of this metabolic trait in this strain. Here, we investigated this instability and describe the finding of a low-abundance and slow-growing actinobacterium, thriving only in co-culture with strain PR1. This organism, named GP, shared highest 16S rRNA gene sequence similarity (94.6–96.9%) with the type strains of validly described species of the genus Leucobacter. This microbial consortium was found to harbor a homolog to the sulfonamide monooxygenase gene (sadA) also found in other sulfonamide-degrading bacteria. This gene is overexpressed in the presence of the antibiotic, and evidence suggests that it codes for a group D flavin monooxygenase responsible for the ipso-hydroxylation of SMX. Additional side reactions were also detected comprising an NIH shift and a Baeyer–Villiger rearrangement, which indicate an inefficient biological transformation of these antibiotics in the environment. This work contributes to further our knowledge in the degradation of this ubiquitous micropollutant by environmental bacteria.






Similar content being viewed by others
References
Baeyer A, Villiger V (1899) Einwirkung des caro’schen reagens auf ketone. Berichte der Dtsch Chem Gesellschaft 32:3625–3633. https://doi.org/10.1002/cber.189903203151
Barreiros L, Nogales B, Manaia CM, Silva Ferreira AC, Pieper DH, Reis MA, Nunes OC (2003) A novel pathway for mineralization of the thiocarbamate herbicide molinate by a defined bacterial mixed culture. Environ Microbiol 5:944–953. https://doi.org/10.1046/j.1462-2920.2003.00492.x
Bhuiyan MNI, Takai R, Mitsuhashi S, Shigetomi K, Tanaka Y, Kamagata Y, Ubukata M (2015) Zincmethylphyrins and coproporphyrins, novel growth factors released by Sphingopyxis sp., enable laboratory cultivation of previously uncultured Leucobacter sp. through interspecies mutualism. J Antibiot (Tokyo) 69:97–103. https://doi.org/10.1038/ja.2015.87
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523
Bouju H, Ricken B, Beffa T, Corvini PFX, Kolvenbach BA (2012) Isolation of bacterial strains capable of sulfamethoxazole mineralization from an acclimated membrane bioreactor. Appl Environ Microbiol 78:277–279. https://doi.org/10.1128/AEM.05888-11
Brankatschk R, Bodenhausen N, Zeyer J, Bürgmann H (2012) Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl Environ Microbiol 78:4481–4489. https://doi.org/10.1128/AEM.07878-11
Chen J, Xie S (2018) Overview of sulfonamide biodegradation and the relevant pathways and microorganisms. Sci Total Environ 640–641:1465–1477. https://doi.org/10.1016/j.scitotenv.2018.06.016
Chen Y, Zhang H, Luo Y, Song J (2012) Occurrence and assessment of veterinary antibiotics in swine manures: a case study in East China. Chin Sci Bull 57:606–614. https://doi.org/10.1007/s11434-011-4830-3
Cribb AE, Spielberg SP (1992) Sulfamethoxazole is metabolized to the hydroxylamine in humans. Clin Pharmacol Ther 51:522–526. https://doi.org/10.1038/clpt.1992.57
Deng Y, Mao Y, Li B, Yang C, Zhang T (2016) Aerobic degradation of sulfadiazine by Arthrobacter spp.: kinetics, pathways, and genomic characterization. Environ Sci Technol 50:9566–9575. https://doi.org/10.1021/acs.est.6b02231
Deng Y, Li B, Zhang T (2018) Bacteria that make a meal of sulfonamide antibiotics: blind spots and emerging opportunities. Environ Sci Technol 52:3854–3868. https://doi.org/10.1021/acs.est.7b06026
El-Awaad I, Bocola M, Beuerle T, Liu B, Beerhues L (2016) Bifunctional CYP81AA proteins catalyse identical hydroxylations but alternative regioselective phenol couplings in plant xanthone biosynthesis. Nat Commun 7:11472. https://doi.org/10.1038/ncomms11472
European Medicines Agency - EMA (2016) Sales of veterinary antimicrobial agents in 29 European countries in 2014. European surveillance of veterinary antimicrobial consumption (EMA/61769/2016)
Gabriel FLP, Heidlberger A, Rentsch D, Giger W, Guenther K, Kohler H-PE (2005) A novel metabolic pathway for degradation of 4-nonylphenol environmental contaminants by Sphingomonas xenophaga Bayram: ipso-hydroxylation and intramolecular rearrangement. J Biol Chem 280:15526–15533. https://doi.org/10.1074/jbc.M413446200
García-Galán MJ, Silvia Díaz-Cruz M, Barceló D (2008) Identification and determination of metabolites and degradation products of sulfonamide antibiotics. TrAC Trends Anal Chem 27:1008–1022. https://doi.org/10.1016/j.trac.2008.10.001
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788. https://doi.org/10.1093/nar/gkg563
Gorsky LD, Koop DR, Coon MJ (1984) On the stoichiometry of the oxidase and monooxygenase reactions catalyzed by liver microsomal cytochrome P-450. Products of oxygen reduction. J Biol Chem 259:6812–6817
Guroff G, Renson J, Udenfriend S, Daly JW, Jerina DM, Witkop B (1967) Hydroxylation-induced migration: the NIH shift. Science 157:1524–1530
Hao X, Jiang R, Chen T (2011) Clustering 16S rRNA for OTU prediction: a method of unsupervised Bayesian clustering. Bioinformatics 27:611–618. https://doi.org/10.1093/bioinformatics/btq725
Hays SG, Patrick WG, Ziesack M, Oxman N, Silver PA (2015) Better together: engineering and application of microbial symbioses. Curr Opin Biotechnol 36:40–49. https://doi.org/10.1016/j.copbio.2015.08.008
Heuer H, Schmitt H, Smalla K (2011) Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol 14:236–243. https://doi.org/10.1016/j.mip.2011.04.009
Hillas PJ, Fitzpatrick PF (1996) A mechanism for hydroxylation by tyrosine hydroxylase based on partitioning of substituted phenylalanines. Biochemistry 35:6969–6975. https://doi.org/10.1021/bi9606861
Hu L, Flanders PM, Miller PL, Strathmann TJ (2007) Oxidation of sulfamethoxazole and related antimicrobial agents by TiO2 photocatalysis. Water Res 41:2612–2626. https://doi.org/10.1016/j.watres.2007.02.026
Huijbers MME, Montersino S, Westphal AH, Tischler D, van Berkel WJH (2014) Flavin dependent monooxygenases. Arch Biochem Biophys 544:2–17. https://doi.org/10.1016/j.abb.2013.12.005
Ingerslev F, Halling-Sørensen B (2000) Biodegradability properties of sulfonamides in activated sludge. Environ Toxicol Chem 19:2467–2473. https://doi.org/10.1002/etc.5620191011
Islas-Espinoza M, Reid BJ, Wexler M, Bond PL (2012) Soil bacterial consortia and previous exposure enhance the biodegradation of sulfonamides from pig manure. Microb Ecol 64:140–151. https://doi.org/10.1007/s00248-012-0010-5
Jiang B, Li A, Cui D, Cai R, Ma F, Wang Y (2014) Biodegradation and metabolic pathway of sulfamethoxazole by Pseudomonas psychrophila HA-4, a newly isolated cold-adapted sulfamethoxazole-degrading bacterium. Appl Microbiol Biotechnol 98:4671–4681. https://doi.org/10.1007/s00253-013-5488-3
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. https://doi.org/10.1093/nar/gkn201
Kneifel H, Elmendorff K, Hegewald E, Soeder CJ (1997) Biotransformation of 1-naphthalenesulfonic acid by the green alga Scenedesmus obliquus. Arch Microbiol 167:32–37. https://doi.org/10.1007/s002030050413
Kolvenbach B, Schlaich N, Raoui Z, Prell J, Zühlke S, Schäffer A, Guengerich FP, Corvini PFX (2007) Degradation pathway of bisphenol A: ipso substitution apply to phenols containing a quaternary alpha-carbon structure in the para position? Appl Environ Microbiol 73:4776–4784. https://doi.org/10.1128/aem.00329-07
Kolvenbach BA, Dobrowinski H, Fousek J, Vlcek C, Schäffer A, Gabriel FLP, Kohler H-PE, Corvini PFX (2012) An unexpected gene cluster for downstream degradation of alkylphenols in Sphingomonas sp. strain TTNP3. Appl Microbiol Biotechnol 93:1315–1324. https://doi.org/10.1007/s00253-011-3451-8
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. https://doi.org/10.1101/gr.215087.116
Krom MD (1980) Spectrophotometric determination of ammonia: a study of a modified Berthelot reaction using salicylate and dichloroisocyanurate. Analyst 105:305–316. https://doi.org/10.1039/an9800500305
Kümmerer K (2009) Antibiotics in the aquatic environment—a review—part I. Chemosphere 75:417–434. https://doi.org/10.1016/j.chemosphere.2008.11.086
Lane DJ (1991) 16S/23S rRNA sequencing. John Wiley and Sons Ltd., New York
Larcher S, Yargeau V (2012) Biodegradation of sulfamethoxazole: current knowledge and perspectives. Appl Microbiol Biotechnol 96:309–318. https://doi.org/10.1007/s00253-012-4326-3
Laskov C, Herzog C, Lewandowski J, Hupfer M (2007) Miniaturized photometrical methods for the rapid analysis of phosphate, ammonium, ferrous iron, and sulfate in pore water of freshwater sediments. Limnol Oceanogr Methods 5:63–71. https://doi.org/10.4319/lom.2007.5.63
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lykidis A, Chen C-L, Tringe SG, McHardy AC, Copeland A, Kyrpides NC, Hugenholtz P, Macarie H, Olmos A, Monroy O, Liu W-T (2011) Multiple syntrophic interactions in a terephthalate-degrading methanogenic consortium. ISME J 5:122–130. https://doi.org/10.1038/ismej.2010.125
Manaia CM, Macedo G, Fatta-Kassinos D, Nunes OC (2016) Antibiotic resistance in urban aquatic environments: can it be controlled? Appl Microbiol Biotechnol 100:1543–1557. https://doi.org/10.1007/s00253-015-7202-0
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. https://doi.org/10.1093/nar/gkw1129
Martinez JL (2009) Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut 157:2893–2902. https://doi.org/10.1016/j.envpol.2009.05.051
Masters PA, O’Bryan TA, Zurlo J, Miller DQ, Joshi N, P G WES (2003) Trimethoprim–sulfamethoxazole revisited. Arch Intern Med 163:402–410. https://doi.org/10.1001/archinte.163.4.402
Mulla SI, Hu A, Sun Q, Li J, Suanona F, Ashfaq M, Yu C-P (2018) Biodegradation of sulfamethoxazole in bacteria from three different origins. J Environ Manag 206:93–102. https://doi.org/10.1016/j.jenvman.2017.10.029
Müller E, Schüssler W, Horn H, Lemmer H (2013) Aerobic biodegradation of the sulfonamide antibiotic sulfamethoxazole by activated sludge applied as co-substrate and sole carbon and nitrogen source. Chemosphere 92:969–978. https://doi.org/10.1016/j.chemosphere.2013.02.070
Owens N (2012) NIH shift literature search. In: All Capstone Proj. http://opus.govst.edu/capstones/72. Accessed 18 Oct 2016
Pelz O, Tesar M, Wittich R-M, Moore ERB, Timmis KN, Abraham W-R (1999) Towards elucidation of microbial community metabolic pathways: unravelling the network of carbon sharing in a pollutant-degrading bacterial consortium by immunocapture and isotopic ratio mass spectrometry. Environ Microbiol 1:167–174. https://doi.org/10.1046/j.1462-2920.1999.00023.x
Pérez S, Eichhorn P, Aga DS (2005) Evaluating the biodegradability of sulfamethazine, sulfamethoxazole, sulfathiazole, and trimethoprim at different stages of sewage treatment. Environ Toxicol Chem 24:1361–1367. https://doi.org/10.1897/04-211R.1
Perreten V, Boerlin P (2003) A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother 47:1169–1172. https://doi.org/10.1128/aac.47.3.1169-1172.2003
Razavi M, Marathe NP, Gillings MR, Flach C-F, Kristiansson E, Joakim Larsson DG (2017) Discovery of the fourth mobile sulfonamide resistance gene. Microbiome 5:160–172. https://doi.org/10.1186/s40168-017-0379-y
Reis PJM, Reis AC, Ricken B, Kolvenbach BA, Manaia CM, Corvini PFX, Nunes OC (2014) Biodegradation of sulfamethoxazole and other sulfonamides by Achromobacter denitrificans PR1. J Hazard Mater 280:741–749. https://doi.org/10.1016/j.jhazmat.2014.08.039
Reis AC, Kroll K, Gomila M, Kolvenbach BA, Corvini PFX, Nunes OC (2017) Complete genome sequence of Achromobacter denitrificans PR1. Genome Announc 5:e00762–e00717. https://doi.org/10.1128/genomeA.00762-17
Reis PJM, Homem V, Alves A, Vilar VJP, Manaia CM, Nunes OC (2018) Insights on sulfamethoxazole bio-transformation by environmental Proteobacteria isolates. J Hazard Mater 358:310–318. https://doi.org/10.1016/j.jhazmat.2018.07.012
Ricken B, Corvini PFX, Cichocka D, Parisi M, Lenz M, Wyss D, Martínez-Lavanchy PM, Müller JA, Shahgaldian P, Tulli LG, Kohler H-PE, Kolvenbach BA (2013) Ipso-hydroxylation and subsequent fragmentation: a novel microbial strategy to eliminate sulfonamide antibiotics. Appl Environ Microbiol 79:5550–5558. https://doi.org/10.1128/aem.00911-13
Ricken B, Fellmann O, Kohler H-PE, Schäffer A, Corvini PF-X, Kolvenbach BA (2015a) Degradation of sulfonamide antibiotics by Microbacterium sp. strain BR1—elucidating the downstream pathway. New Biotechnol 32:710–715. https://doi.org/10.1016/j.nbt.2015.03.005
Ricken B, Kolvenbach BA, Corvini PF-X (2015b) Ipso-substitution—the hidden gate to xenobiotic degradation pathways. Curr Opin Biotechnol 33:220–227. https://doi.org/10.1016/j.copbio.2015.03.009
Ricken B, Kolvenbach BA, Bergesch C, Benndorf D, Kroll K, Strnad H, Vlček Č, Adaixo R, Hammes F, Shahgaldian P, Schäffer A, Kohler H-PE, Corvini PF-X (2017) FMNH2-dependent monooxygenases initiate catabolism of sulfonamides in Microbacterium sp. strain BR1 subsisting on sulfonamide antibiotics. Sci Rep 7:15783. https://doi.org/10.1038/s41598-017-16132-8
Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D (2013) Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. https://doi.org/10.1016/j.scitotenv.2013.01.032
Sanderson JP, Hollis FJ, Maggs JL, Clarke SE, Naisbitt DJ, Park BK (2008) Nonenzymatic formation of a novel hydroxylated sulfamethoxazole derivative in human liver microsomes: implications for bioanalysis of sulfamethoxazole metabolites. Drug Metab Dispos 36:2424–2428. https://doi.org/10.1124/dmd.108.021246
Sarmah AK, Meyer MT, Boxall ABA (2006) A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65:725–759. https://doi.org/10.1016/j.chemosphere.2006.03.026
Schink B (2002) Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81:257–261. https://doi.org/10.1023/a:1020579004534
Sköld O (2000) Sulfonamide resistance: mechanisms and trends. Drug Resist Updat 3:155–160. https://doi.org/10.1054/drup.2000.0146
Sørensen SR, Ronen Z, Aamand J (2002) Growth in coculture stimulates metabolism of the phenylurea herbicide isoproturon by Sphingomonas sp. strain SRS2. Appl Environ Microbiol 68:3478–3485. https://doi.org/10.1128/aem.68.7.3478-3485.2002
Su T, Deng H, Benskin JP, Radke M (2016) Biodegradation of sulfamethoxazole photo-transformation products in a water/sediment test. Chemosphere 148:518–525. https://doi.org/10.1016/j.chemosphere.2016.01.049
Takenaka S, Okugawa S, Kadowaki M, Murakami S, Aoki K (2003) The metabolic pathway of 4-aminophenol in Burkholderia sp. strain AK-5 differs from that of aniline and aniline with C-4 substituents. Appl Environ Microbiol 69:5410–5413. https://doi.org/10.1128/aem.69.9.5410-5413.2003
Tappe W, Herbst M, Hofmann D, Koeppchen S, Kummer S, Thiele B, Groeneweg J (2013) Degradation of sulfadiazine by Microbacterium lacus strain SDZm4, isolated from lysimeters previously manured with slurry from sulfadiazine-medicated pigs. Appl Environ Microbiol 79:2572–2577. https://doi.org/10.1128/aem.03636-12
Taupp M, Heckel F, Harmsen D, Schreier P (2006) Biohydroxylation of N,N-dialkylarylamines by the isolated topsoil bacterium Bacillus megaterium. Enzym Microb Technol 38:1013–1016. https://doi.org/10.1016/j.enzmictec.2005.11.021
Teuber M (2001) Veterinary use and antibiotic resistance. Curr Opin Microbiol 4:493–499. https://doi.org/10.1016/s1369-5274(00)00241-1
Topp E, Chapman R, Devers-Lamrani M, Hartmann A, Marti R, Martin-Laurent F, Sabourin L, Scott A, Sumarah M (2012) Accelerated biodegradation of veterinary antibiotics in agricultural soil following long-term exposure, and isolation of a sulfamethazine-degrading Microbacterium sp. J Environ Qual 42:173–178. https://doi.org/10.2134/jeq2012.0162
Turner S, Pryer KM, Miao VPW, Palmer JD (1999) Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46:327–338. https://doi.org/10.1111/j.1550-7408.1999.tb04612.x
Ullrich R, Hofrichter M (2007) Enzymatic hydroxylation of aromatic compounds. Cell Mol Life Sci 64:271–293. https://doi.org/10.1007/s00018-007-6362-1
Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, Laxminarayan R (2014) Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis 14:742–750. https://doi.org/10.1016/s1473-3099(14)70780-7
Vartoukian SR, Adamowska A, Lawlor M, Moazzez R, Dewhirst FE, Wade WG (2016) In vitro cultivation of ‘unculturable’ oral bacteria, facilitated by community culture and media supplementation with siderophores. PLoS One 11:e0146926. https://doi.org/10.1371/journal.pone.0146926
Wada A, Kono M, Kawauchi S, Takagi Y, Morikawa T, Funakoshi K (2012) Rapid discrimination of gram-positive and gram-negative bacteria in liquid samples by using NaOH-sodium dodecyl sulfate solution and flow cytometry. PLoS One 7:e47093. https://doi.org/10.1371/journal.pone.0047093
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/aem.00062-07
Wang S, Hu Y, Wang J (2018) Biodegradation of typical pharmaceutical compounds by a novel strain Acinetobacter sp. J Environ Manag 217:240–246. https://doi.org/10.1016/j.jenvman.2018.03.096
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Wintermute EH, Silver PA (2010) Dynamics in the mixed microbial concourse. Genes Dev 24:2603–2614. https://doi.org/10.1101/gad.1985210
Wolf S, Schmidt S, Müller-Hannemann M, Neumann S (2010) In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics 11:148. https://doi.org/10.1186/1471-2105-11-148
Wright ES, Yilmaz LS, Noguera DR (2012) DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78:717–725. https://doi.org/10.1128/aem.06516-11
Wu J-H, Wu F-Y, Chuang H-P, Chen W-Y, Huang H-J, Chen S-H, Liu W-T (2013) Community and proteomic analysis of methanogenic consortia degrading terephthalate. Appl Environ Microbiol 79:105–112. https://doi.org/10.1128/aem.02327-12
Yagi H, Jerina DM, Kasperek GJ, Bruice TC (1972) A novel mechanism for the NIH-shift. Proc Natl Acad Sci U S A 69:1985–1986
Yang N, Wan J, Zhao S, Wang Y (2015) Removal of concentrated sulfamethazine by acclimatized aerobic sludge and possible metabolic products. PeerJ 3:e1359. https://doi.org/10.7717/peerj.1359
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. https://doi.org/10.1186/1471-2105-13-134
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Zakharieva O, Grodzicki M, Trautwein AX, Veeger C, Rietjens IMC (1998) Molecular orbital study of porphyrin–substrate interactions in cytochrome P450 catalysed aromatic hydroxylation of substituted anilines. Biophys Chem 73:189–203. https://doi.org/10.1016/s0301-4622(98)00111-2
Zhang Y-B, Zhou J, Xu Q-M, Cheng J-S, Luo Y-L, Yuan Y-J (2016) Exogenous cofactors for the improvement of bioremoval and biotransformation of sulfamethoxazole by Alcaligenes faecalis. Sci Total Environ 565:547–556. https://doi.org/10.1016/j.scitotenv.2016.05.063
Acknowledgments
The authors wish to thank Anna Weston (FHNW, Switzerland) for the advice on the optimization of the qPCR assays, Jonas Romer (FHNW, Switzerland) for the assistance with the TiO2/UV experiments, Prof. Dr. Hans-Peter Kohler (EWAG, Switzerland) for the helpful insights about the metabolic pathway, and Dr. Ana Rita Lopes (FEUP, Portugal) for the relevant comments about the structure of the manuscript and presentation of results. The authors wish to acknowledge the Swiss National Science Foundation, Comissão de Coordenação e Desenvolvimento Regional do Norte and Fundação para a Ciência e a Tecnologia for the funding. Ana Reis further acknowledges the Fundação para a Ciência e a Tecnologia (FCT) for her PhD scholarship.
Funding
This work was financially supported by the Swiss National Science Foundation (grant no. 160332), Comissão de Coordenação e Desenvolvimento Regional do Norte (project reference NORTE-01-0145-FEDER-000005), and Fundação para a Ciência e a Tecnologia and Technology (FCT) through the project UID/EQU/00511/2013—POCI-01-0145-FEDER-006939 and Ana Reis PhD grant (reference SFRH/BD/95814/2013).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 1657 kb)
Rights and permissions
About this article
Cite this article
Reis, A.C., Čvančarová, M., Liu, Y. et al. Biodegradation of sulfamethoxazole by a bacterial consortium of Achromobacter denitrificans PR1 and Leucobacter sp. GP. Appl Microbiol Biotechnol 102, 10299–10314 (2018). https://doi.org/10.1007/s00253-018-9411-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9411-9