Abstract
Purpose
Attenuation correction using CT transmission scanning increases the accuracy of single-photon emission computed tomography (SPECT) and enables quantitative analysis. Current existing SPECT-only systems normally do not support transmission scanning and therefore scans on these systems are susceptible to attenuation artifacts. Moreover, the use of CT scans also increases radiation dose to patients and significant artifacts can occur due to the misregistration between the SPECT and CT scans as a result of patient motion. The purpose of this study is to develop an approach to estimate attenuation maps directly from SPECT emission data using deep learning methods.
Methods
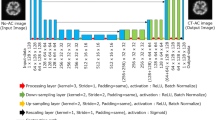
Both photopeak window and scatter window SPECT images were used as inputs to better utilize the underlying attenuation information embedded in the emission data. The CT-based attenuation maps were used as labels with which cardiac SPECT/CT images of 65 patients were included for training and testing. We implemented and evaluated deep fully convolutional neural networks using both standard training and training using an adversarial strategy.
Results
The synthetic attenuation maps were qualitatively and quantitatively consistent with the CT-based attenuation map. The globally normalized mean absolute error (NMAE) between the synthetic and CT-based attenuation maps were 3.60% ± 0.85% among the 25 testing subjects. The SPECT reconstructed images corrected using the CT-based attenuation map and synthetic attenuation map are highly consistent. The NMAE between the reconstructed SPECT images that were corrected using the synthetic and CT-based attenuation maps was 0.26% ± 0.15%, whereas the localized absolute percentage error was 1.33% ± 3.80% in the left ventricle (LV) myocardium and 1.07% ± 2.58% in the LV blood pool.
Conclusion
We developed a deep convolutional neural network to estimate attenuation maps for SPECT directly from the emission data. The proposed method is capable of generating highly reliable attenuation maps to facilitate attenuation correction for SPECT-only scanners for myocardial perfusion imaging.







Similar content being viewed by others
References
van Dijk J, Mouden M, Ottervanger J, van Dalen J, Knollema S, Slump C, et al. Value of attenuation correction in stress-only myocardial perfusion imaging using CZT-SPECT. J Nucl Cardiol. 2017;24:395–401.
Patchett ND, Pawar S, Sverdlov A, Miller EJ. Does Improved Technology in SPECT Myocardial Perfusion Imaging Reduce Downstream Costs? An Observational Study. International Journal of Radiology and Imaging Technology. 2017;3. https://doi.org/10.23937/2572-3235.1510023.
Abdollahi H, Shiri I, Salimi Y, Sarebani M, Mehdinia R, Deevband MR, et al. Radiation dose in cardiac SPECT/CT: an estimation of SSDE and effective dose. Eur J Radiol. 2016;85:2257–61.
Technavio. Global SPECT Market 2017-2021. 2017. https://www.technavio.com/report/global-medical-imaging-global-spect-market-2017-2021. Accessed 5 Nov 2019.
Jha AK, Zhu Y, Clarkson E, Kupinski MA, Frey EC. Fisher information analysis of list-mode SPECT emission data for joint estimation of activity and attenuation distribution. arXiv preprint. arXiv:180701767. 2018.
Wu J, Liu C. Recent advances in cardiac SPECT instrumentation and imaging methods. Phys Med Biol. 2019;64:06TR1.
Shimizu M, Fujii H, Yamawake N, Nishizaki M. Cardiac function changes with switching from the supine to prone position: analysis by quantitative semiconductor gated single-photon emission computed tomography. J Nucl Cardiol. 2015;22:301–7.
Pan T-S, King MA, Luo D-S, Dahlberg ST, Villegas BJ. Estimation of attenuation maps from scatter and photopeak window single photon-emission computed tomographic images of technetium 99m-labeled sestamibi. J Nucl Cardiol. 1997;4:42–51.
Zaidi H, Hasegawa B. Determination of the attenuation map in emission tomography. J Nucl Med. 2003;44:291–315.
Núñez M, Prakash V, Vila R, Mut F, Alonso O, Hutton BF. Attenuation correction for lung SPECT: evidence of need and validation of an attenuation map derived from the emission data. Eur J Nucl Med Mol Imaging. 2009;36:1076–89.
Pan T-S, King MA, de Vries DJ, Ljungberg M. Segmentation of the body and lungs from Compton scatter and photopeak window data in SPECT: a Monte-Carlo investigation. IEEE Trans Med Imaging. 1996;15:13–24.
Cade SC, Arridge S, Evans MJ, Hutton BF. Use of measured scatter data for the attenuation correction of single photon emission tomography without transmission scanning. Med Phys. 2013;40:082506.
Gourion D, Noll D, Gantet P, Celler A, Esquerré J-P. Attenuation correction using SPECT emission data only. IEEE Trans Nucl Sci. 2002;49:2172–9.
Yan Y, Zeng GL. Attenuation map estimation with SPECT emission data only. Int J Imaging Syst Technol. 2009;19:271–6.
Nuyts J, Dupont P, Stroobants S, Benninck R, Mortelmans L, Suetens P. Simultaneous maximum a posteriori reconstruction of attenuation and activity distributions from emission sinograms. IEEE Trans Med Imaging. 1999;18:393–403.
Krol A, Bowsher JE, Manglos SH, Feiglin DH, Tornai MP, Thomas FD. An EM algorithm for estimating SPECT emission and transmission parameters from emission data only. IEEE Trans Med Imaging. 2001;20:218–32.
Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, et al. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans Biomed Eng. 2018;65:2720–30.
Hwang D, Kang SK, Kim KY, Seo S, Paeng JC, Lee DS, et al. Generation of PET attenuation map for whole-body time-of-flight 18F-FDG PET/MRI using a deep neural network trained with simultaneously reconstructed activity and attenuation maps. J Nucl Med. 2019:jnumed. 118.219493.
Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys. 2017;44:1408–19.
Shi L, Onofrey JA, Revilla EM, Toyonaga T, Menard D, Ankrah J, et al. A novel loss function incorporating imaging acquisition physics for PET attenuation map generation using deep learning. International Conference on Medical image computing and computer-assisted intervention: Springer International Publishing; 2019. p. 723–31.
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Advances in neural information processing systems; 2014. p. 2672–80.
Mao X, Li Q, Xie H, Lau RY, Wang Z, Paul Smolley S. Least squares generative adversarial networks. Proceedings of the IEEE International Conference on Computer Vision; 2017. p. 2794–802.
Shi L, Onofrey J, Liu H, Liu Y-H, Liu C. Generating attenuation map for SPECT-only systems using generative adversarial networks. J Nucl Med. 2019;60:572.
Hudson HM, Larkin RS. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans Med Imaging. 1994;13:601–9.
Isola P, Zhu J-Y, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 1125–34.
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention: Springer; 2015. p. 234–41.
Milletari F, Navab N, Ahmadi S-A. V-net: fully convolutional neural networks for volumetric medical image segmentation. 2016 Fourth International Conference on 3D Vision (3DV): IEEE; 2016. p. 565–71.
Onofrey JA, Casetti-Dinescu DI, Lauritzen AD, Sarkar S, Venkataraman R, Fan RE, et al. Generalizable multi-site training and testing of deep neural networks using image normalization. Biomedical Imaging (ISBI), 2019 IEEE 16th International Symposium on; 2019. p. pp. 1–4.
Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. Tensorflow: a system for large-scale machine learning. 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16); 2016. p. 265–83.
Acknowledgments
This study was supported by American Heart Association award 18PRE33990138 and National Institute of Health grant R01HL123949. We would like to thank Dr. Eric Frey from Johns Hopkins University for the image reconstruction tool and Dr. Stephanie Thorn from Yale University and Christopher Weyman from Yale New Haven Hospital for helpful discussions. We would also like to thank Drs. Edward J. Miller and Albert J. Sinusas from Yale University for their help with data acquisition and results interpretation.
Funding
This study was supported by American Heart Association award 18PRE33990138 and National Institute of Health grant R01HL123949.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Conception and design of the study: Luyao Shi and Chi Liu; algorithm implementation: Luyao Shi and John A. Onofrey; data acquisition: Luyao Shi and Chi Liu; data analysis: Luyao Shi, Hui Liu, and Yi-Hwa Liu; writing of the first draft of the manuscript: Luyao Shi. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Chi Liu, Luyao Shi, and John Onofrey are named inventors on a provisional patent application that Yale University has filed on this work. No other potential conflicts of interest relevant to this article exist.
Ethical approval retrospective studies
The retrospective use of the anonymized data in this study was approved by Yale Institutional Review Board under protocol number 2000026790.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Cardiology
Electronic supplementary material
ESM 1
(DOCX 33 kb)
Rights and permissions
About this article
Cite this article
Shi, L., Onofrey, J.A., Liu, H. et al. Deep learning-based attenuation map generation for myocardial perfusion SPECT. Eur J Nucl Med Mol Imaging 47, 2383–2395 (2020). https://doi.org/10.1007/s00259-020-04746-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-020-04746-6