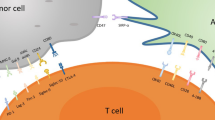
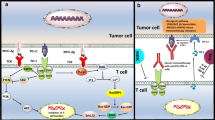
Abstract
Immunotherapy aims to activate the immune system to fight cancer in a very specific and targeted manner. Despite the success of different immunotherapeutic strategies, in particular antibodies directed against checkpoints as well as adoptive T-cell therapy, the response of patients is limited in different types of cancers. This attributes to escape of the tumor from immune surveillance and development of acquired resistances during therapy. In this review, the different evasion and resistance mechanisms that limit the efficacy of immunotherapies targeting tumor-associated antigens presented by major histocompatibility complex molecules on the surface of the malignant cells are summarized. Overcoming these escape mechanisms is a great challenge, but might lead to a better clinical outcome of patients and is therefore currently a major focus of research.


Similar content being viewed by others
Abbreviations
- DAC:
-
5-Aza-2′-desoxycytidine
- 3′ UTR:
-
3′ Untranslated region
- ACT:
-
Adoptive cell therapy
- APM:
-
Antigen processing and presentation machinery
- BGN:
-
Biglycan
- CTL:
-
Cytotoxic T lymphocyte
- CTLA-4:
-
Cytotoxic T-lymphocyte associated protein-4
- DCN:
-
Decorin
- ER:
-
Endoplasmic reticulum
- ERAP:
-
Endoplasmic reticulum aminopeptidase
- EBV:
-
Epstein–Barr virus
- GAS:
-
Gamma activated site
- HC:
-
Heavy chain
- HNRNPR:
-
Heterogeneous nuclear ribonucleoprotein R
- HCMV:
-
Human cytomegalovirus
- iCP:
-
Immune checkpoint
- iCPI:
-
Immune checkpoint inhibitor
- im-miRNAs:
-
Immune modulatory miRNAs
- ILT:
-
Inhibitory receptor Ig-like transcript
- IFN:
-
Interferon
- IRF:
-
Interferon regulated factor
- ISRE:
-
Interferon-sensitive response element
- LOH:
-
Loss of heterozygosity
- Luc:
-
Luciferase
- MHC:
-
Major histocompatibility complex
- β2-m:
-
β2-Microglobulin
- miRNA:
-
MicroRNA
- MSI:
-
Microsatellite instability
- Mex:
-
Muscle excess
- MDSC:
-
Myeloid-derived suppressor cells
- NLRC5:
-
NOD-like receptor family and acid domain-containing protein 5
- OS:
-
Overall survival
- Treg:
-
Regulatory T cell(s)
- RBP:
-
RNA-binding proteins
- RNAseq:
-
RNA sequencing
- STAT:
-
Signal transducer and activator of transcription
- SNP:
-
Single nucleotide polymorphism
- SLRP:
-
Small leucine-rich proteoglycan
- TPN:
-
Tapasin
- TCGA:
-
The Cancer Genome Atlas
- TGF-β:
-
Transforming growth factor β
- TAP:
-
Transporter associated with antigen processing
- TAF:
-
Tumor-associated fibroblast(s)
- TAM:
-
Tumor-associated macrophages
- TAN:
-
Tumor-associated neutrophil(s)
- TMB:
-
Tumor mutational burden
- TME:
-
Tumor microenvironment
References
Leone P et al (2013) MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J Natl Cancer Inst 105(16):1172–1187
Zanker D, Chen W (2014) Standard and immunoproteasomes show similar peptide degradation specificities. Eur J Immunol 44(12):3500–3503
Saveanu L et al (2005) Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol 6(7):689–697
Norbury CC, Eisenlohr LC (2016) Editorial overview: antigen processing. Curr Opin Immunol 40:5–6
Mintern JD, Macri C, Villadangos JA (2015) Modulation of antigen presentation by intracellular trafficking. Curr Opin Immunol 34:16–21
Han LY et al (2008) HLA class I antigen processing machinery component expression and intratumoral T-Cell infiltrate as independent prognostic markers in ovarian carcinoma. Clin Cancer Res 14(11):3372–3379
Perea F et al (2017) The absence of HLA class I expression in non-small cell lung cancer correlates with the tumor tissue structure and the pattern of T cell infiltration. Int J Cancer 140(4):888–899
Garrido F et al (2017) The escape of cancer from T cell-mediated immune surveillance: HLA class I loss and tumor tissue architecture. Vaccines (Basel) 5(1):7
Seliger B (2016) Role of microRNAs on HLA-G expression in human tumors. Hum Immunol 77(9):760–763
Real LM et al (1999) Expression of HLA G in human tumors is not a frequent event. Int J Cancer 81(4):512–518
Davies B et al (2001) HLA-G expression by tumors. Am J Reprod Immunol 45(2):103–107
Polakova K et al (2003) Analysis of HLA-G expression in malignant hematopoietic cells from leukemia patients. Leuk Res 27(7):643–648
Frumento G et al (2000) Melanomas and melanoma cell lines do not express HLA-G, and the expression cannot be induced by gamma IFN treatment. Tissue Antigens 56(1):30–37
Pangault C et al (1999) HLA-G protein expression is not induced during malignant transformation. Tissue Antigens 53(4 Pt 1):335–346
Chang CC, Ferrone S (2003) HLA-G in melanoma: can the current controversies be solved? Semin Cancer Biol 13(5):361–369
Wang Y et al (2011) Expression of HLA-G in patients with hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int 10(2):158–163
Yie SM et al (2007) Expression of human leucocyte antigen G (HLA-G) is associated with prognosis in non-small cell lung cancer. Lung Cancer 58(2):267–274
Tronik-Le Roux D et al (2017) Novel landscape of HLA-G isoforms expressed in clear cell renal cell carcinoma patients. Mol Oncol 11(11):1561–1578
Lin A, Yan WH (2018) Heterogeneity of HLA-G expression in cancers: facing the challenges. Front Immunol 9:2164
Lin A, Yan WH (2015) Human leukocyte antigen-G (HLA-G) expression in cancers: roles in immune evasion, metastasis and target for therapy. Mol Med 21(1):782–791
Contini P et al (2003) Soluble HLA-A,-B,-C and -G molecules induce apoptosis in T and NK CD8 + cells and inhibit cytotoxic T cell activity through CD8 ligation. Eur J Immunol 33(1):125–134
Menier C et al (2002) MICA triggering signal for NK cell tumor lysis is counteracted by HLA-G1-mediated inhibitory signal. Int J Cancer 100(1):63–70
Fons P et al (2006) Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood 108(8):2608–2615
Morandi F et al (2010) A novel mechanism of soluble HLA-G mediated immune modulation: downregulation of T cell chemokine receptor expression and impairment of chemotaxis. PLoS One 5(7):e11763
Morandi F et al (2011) Soluble HLA-G dampens CD94/NKG2A expression and function and differentially modulates chemotaxis and cytokine and chemokine secretion in CD56bright and CD56dim NK cells. Blood 118(22):5840–5850
Agaugue S, Carosella ED, Rouas-Freiss N (2011) Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood 117(26):7021–7031
Loumagne L et al (2014) In vivo evidence that secretion of HLA-G by immunogenic tumor cells allows their evasion from immunosurveillance. Int J Cancer 135(9):2107–2117
Seliger B (2014) The link between MHC class I abnormalities of tumors, oncogenes, tumor suppressor genes, and transcription factors. J Immunotoxicol 11(4):308–310
Cai L et al (2018) Defective HLA class I antigen processing machinery in cancer. Cancer Immunol Immunother 67(6):999–1009
Donia M et al (2017) Acquired Immune resistance follows complete tumor regression without loss of target antigens or IFN gamma signaling. Cancer Res 77(17):4562–4566
Kloor M, Michel S, von Knebel Doeberitz M (2010) Immune evasion of microsatellite unstable colorectal cancers. Int J Cancer 127(5):1001–1010
Hicklin DJ et al (1998) Beta2-Microglobulin mutations, HLA class I antigen loss, and tumor progression in melanoma. J Clin Invest 101(12):2720–2729
Chang CC et al (2005) Immune selection of hot-spot beta 2-microglobulin gene mutations, HLA-A2 allospecificity loss, and antigen-processing machinery component down-regulation in melanoma cells derived from recurrent metastases following immunotherapy. J Immunol 174(3):1462–1471
Geertsen R et al (2002) Loss of single HLA class I allospecificities in melanoma cells due to selective genomic abbreviations. Int J Cancer 99(1):82–87
del Campo AB et al (2014) Immune escape of cancer cells with beta2-microglobulin loss over the course of metastatic melanoma. Int J Cancer 134(1):102–113
McGranahan N et al (2017) Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171(6):1259–1271
Zehir A et al (2017) Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23(6):703–713
Seliger B (2017) Immune modulatory microRNAs as a novel mechanism to revert immune escape of tumors. Cytokine Growth Factor Rev 36:49–56
Ritter C et al (2017) Epigenetic priming restores the HLA class-I antigen processing machinery expression in Merkel cell carcinoma. Sci Rep 7(1):2290
Jongsma MLM, Guarda G, Spaapen RM (2017) The regulatory network behind MHC class I expression. Mol Immunol. https://doi.org/10.1016/j.molimm.2017.12.005
Drukker M et al (2002) Characterization of the expression of MHC proteins in human embryonic stem cells. Proc Natl Acad Sci USA 99(15):9864–9869
Vlkova V et al (2014) Epigenetic regulations in the IFNgamma signalling pathway: IFNgamma-mediated MHC class I upregulation on tumour cells is associated with DNA demethylation of antigen-presenting machinery genes. Oncotarget 5(16):6923–6935
Manning J et al (2008) Induction of MHC class I molecule cell surface expression and epigenetic activation of antigen-processing machinery components in a murine model for human papilloma virus 16-associated tumours. Immunology 123(2):218–227
Setiadi AF et al (2008) Epigenetic enhancement of antigen processing and presentation promotes immune recognition of tumors. Cancer Res 68(23):9601–9607
Dunker K et al (2008) Expression and regulation of non-classical HLA-G in renal cell carcinoma. Tissue Antigens 72(2):137–148
Ramsuran V et al (2015) Epigenetic regulation of differential HLA-A allelic expression levels. Hum Mol Genet 24(15):4268–4275
Gustafsson JR et al (2018) DNMT1 regulates expression of MHC class I in post-mitotic neurons. Mol Brain 11(1):36
Khan AN, Gregorie CJ, Tomasi TB (2008) Histone deacetylase inhibitors induce TAP, LMP, Tapasin genes and MHC class I antigen presentation by melanoma cells. Cancer Immunol Immunother 57(5):647–654
Komatsu Y, Hayashi H (1998) Histone deacetylase inhibitors up-regulate the expression of cell surface MHC class-I molecules in B16/BL6 cells. J Antibiot (Tokyo) 51(1):89–91
Robbins GR et al (2012) Regulation of class I major histocompatibility complex (MHC) by nucleotide-binding domain, leucine-rich repeat-containing (NLR) proteins. J Biol Chem 287(29):24294–24303
Moreau P et al (2003) HLA-G gene repression is reversed by demethylation. Proc Natl Acad Sci USA 100(3):1191–1196
Holling TM et al (2009) Genetic and epigenetic control of the major histocompatibility complex class Ib gene HLA-G in trophoblast cell lines. Ann N Y Acad Sci 1173:538–544
van den Elsen PJ (2011) Expression regulation of major histocompatibility complex class I and class II encoding genes. Front Immunol 2:48
van den Elsen PJ et al (1998) Regulation of MHC class I and II gene transcription: differences and similarities. Immunogenetics 48(3):208–221
Howcroft TK et al (2003) Distinct transcriptional pathways regulate basal and activated major histocompatibility complex class I expression. Mol Cell Biol 23(10):3377–3391
Gobin SJ et al (1998) The role of enhancer A in the locus-specific transactivation of classical and nonclassical HLA class I genes by nuclear factor kappa B. J Immunol 161(5):2276–2283
Gobin SJ et al (1999) Transactivation of classical and nonclassical HLA class I genes through the IFN-stimulated response element. J Immunol 163(3):1428–1434
Gobin SJ et al (2001) The MHC-specific enhanceosome and its role in MHC class I and beta(2)-microglobulin gene transactivation. J Immunol 167(9):5175–5184
van den Elsen PJ et al (2004) Transcriptional regulation of antigen presentation. Curr Opin Immunol 16(1):67–75
Gobin SJ et al (1997) Site alpha is crucial for two routes of IFN gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity 6(5):601–611
Bukur J et al (2010) Identification of E2F1 as an important transcription factor for the regulation of tapasin expression. J Biol Chem 285(40):30419–30426
Zheng P et al (1998) Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature 396(6709):373–376
Geiser AG et al (1993) Transforming growth factor beta 1 (TGF-beta 1) controls expression of major histocompatibility genes in the postnatal mouse: aberrant histocompatibility antigen expression in the pathogenesis of the TGF-beta 1 null mouse phenotype. Proc Natl Acad Sci USA 90(21):9944–9948
Baldeon ME et al (1997) Interferon-gamma independently activates the MHC class I antigen processing pathway and diminishes glucose responsiveness in pancreatic beta-cell lines. Diabetes 46(5):770–778
Israel A et al (1989) TNF stimulates expression of mouse MHC class I genes by inducing an NF kappa B-like enhancer binding activity which displaces constitutive factors. EMBO J 8(12):3793–3800
Komov L et al (2018) Cell surface MHC class I expression is limited by the availability of peptide-receptive “empty” molecules rather than by the supply of peptide ligands. Proteomics 18(12):e1700248
Parker BS, Rautela J, Hertzog PJ (2016) Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer 16(3):131–144
Schroder K et al (2004) Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75(2):163–189
Ting JP, Baldwin AS (1993) Regulation of MHC gene expression. Curr Opin Immunol 5(1):8–16
Hertzog PJ, Williams BR (2013) Fine tuning type I interferon responses. Cytokine Growth Factor Rev 24(3):217–225
Zhou F (2009) Molecular mechanisms of IFN-gamma to up-regulate MHC class I antigen processing and presentation. Int Rev Immunol 28(3–4):239–260
Strehl B et al (2005) Interferon-gamma, the functional plasticity of the ubiquitin-proteasome system, and MHC class I antigen processing. Immunol Rev 207:19–30
Meissner TB et al (2010) NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci USA 107(31):13794–13799
Kulski JK et al (2001) Genomic and phylogenetic analysis of the human CD1 and HLA class I multicopy genes. J Mol Evol 53(6):642–650
Gobin SJ, van den Elsen PJ (1999) The regulation of HLA class I expression: is HLA-G the odd one out? Semin Cancer Biol 9(1):55–59
Gobin SJ et al (2002) HLA-G transactivation by cAMP-response element-binding protein (CREB) An alternative transactivation pathway to the conserved major histocompatibility complex (MHC) class I regulatory routes. J Biol Chem 277(42):39525–39531
Flajollet S et al (2009) RREB-1 is a transcriptional repressor of HLA-G. J Immunol 183(11):6948–6959
Eichmuller SB et al (2017) Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst 109(10):1–14
Seliger B (2008) Different regulation of MHC class I antigen processing components in human tumors. J Immunotoxicol 5(4):361–367
Kulkarni S et al (2011) Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature 472(7344):495–498
Tan Z et al (2007) Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet 81(4):829–834
Zhu XM et al (2010) Overexpression of miR-152 leads to reduced expression of human leukocyte antigen-G and increased natural killer cell mediated cytolysis in JEG-3 cells. Am J Obstet Gynecol 202(6):592
Jasinski-Bergner S et al (2016) Identification of novel microRNAs regulating HLA-G expression and investigating their clinical relevance in renal cell carcinoma. Oncotarget 7(18):26866–26878
Friedrich M et al (2017) The role of the miR-148/-152 family in physiology and disease. Eur J Immunol 47(12):2026–2038
Manaster I et al (2012) MiRNA-mediated control of HLA-G expression and function. PLoS ONE 7(3):e33395
Jasinski-Bergner S et al (2015) Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology 4(6):e1008805
Wang Y et al (2017) MicroRNA-152 regulates immune response via targeting B7-H1 in gastric carcinoma. Oncotarget 8(17):28125–28134
Gao F et al (2013) miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun 431(3):610–616
Bartoszewski R et al (2011) The unfolded protein response (UPR)-activated transcription factor X-box-binding protein 1 (XBP1) induces microRNA-346 expression that targets the human antigen peptide transporter 1 (TAP1) mRNA and governs immune regulatory genes. J Biol Chem 286(48):41862–41870
Albanese M et al (2016) Epstein–Barr virus microRNAs reduce immune surveillance by virus-specific CD8 + T cells. Proc Natl Acad Sci USA 113(42):E6467–E6475
Kim S et al (2011) Human cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nat Immunol 12(10):984–991
Huang L et al (2018) The RNA-binding protein MEX3B mediates resistance to cancer immunotherapy by downregulating HLA-A expression. Clin Cancer Res 24(14):3366–3376
Cano F et al (2012) The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. EMBO J 31(17):3596–3606
Reches A et al (2016) HNRNPR regulates the expression of classical and nonclassical MHC class I proteins. J Immunol 196(12):4967–4976
Shankaran V et al (2001) IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410(6832):1107–1111
Kaplan DH et al (1998) Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA 95(13):7556–7561
Ivashkiv LB, Donlin LT (2014) Regulation of type I interferon responses. Nat Rev Immunol 14(1):36–49
Greenlund AC et al (1994) Ligand-induced IFN gamma receptor tyrosine phosphorylation couples the receptor to its signal transduction system (p91). EMBO J 13(7):1591–1600
Decker T et al (1991) Cytoplasmic activation of GAF, an IFN-gamma-regulated DNA-binding factor. EMBO J 10(4):927–932
Chatterjee-Kishore M et al (2000) How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J 19(15):4111–4122
Rettino A, Clarke NM (2013) Genome-wide identification of IRF1 binding sites reveals extensive occupancy at cell death associated genes. J Carcinog Mutagen (Spec Iss Apoptosis). https://doi.org/10.4172/2157-2518.S6-009
Starr R et al (1997) A family of cytokine-inducible inhibitors of signalling. Nature 387(6636):917–921
Castro F et al (2018) Interferon-Gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol 9:847
Seliger B et al (1997) IFN-gamma-mediated coordinated transcriptional regulation of the human TAP-1 and LMP-2 genes in human renal cell carcinoma. Clin Cancer Res 3(4):573–578
Seliger B, Ruiz-Cabello F, Garrido F (2008) IFN inducibility of major histocompatibility antigens in tumors. Adv Cancer Res 101:249–276
Respa A et al (2011) Association of IFN-gamma signal transduction defects with impaired HLA class I antigen processing in melanoma cell lines. Clin Cancer Res 17(9):2668–2678
Aqbi HF et al (2018) IFN-gamma orchestrates tumor elimination, tumor dormancy, tumor escape, and progression. J Leukoc Biol. https://doi.org/10.1002/JLB.5MIR0917-351R
Schaefer L et al (2017) Proteoglycan neofunctions: regulation of inflammation and autophagy in cancer biology. FEBS J 284(1):10–26
Recktenwald CV et al (2008) Altered detoxification status and increased resistance to oxidative stress by K-ras transformation. Cancer Res 68(24):10086–10093
Recktenwald CV et al (2012) HER-2/neu-mediated down-regulation of biglycan associated with altered growth properties. J Biol Chem 287(29):24320–24329
Subbarayan K et al (2018) Biglycan-mediated upregulation of MHC class I expression in HER-2/neu-transformed cells. Oncoimmunology 7(4):e1373233
Subbarayan K, Seliger B (2018) Tumor-dependent effects of proteoglycans and various glycosaminoglycan synthesizing enzymes and sulfotransferases on patients’ outcome. Curr Cancer Drug Targets 19(3):210–221
Yan L, DeMars LC (2012) Dietary supplementation with methylseleninic acid, but not selenomethionine, reduces spontaneous metastasis of Lewis lung carcinoma in mice. Int J Cancer 131(6):1260–1266
Chen YC et al (2013) Dietary selenium supplementation modifies breast tumor growth and metastasis. Int J Cancer 133(9):2054–2064
Labunskyy VM, Hatfield DL, Gladyshev VN (2014) Selenoproteins: molecular pathways and physiological roles. Physiol Rev 94(3):739–777
Lennicke C et al (2017) Modulation of MHC class I surface expression in B16F10 melanoma cells by methylseleninic acid. Oncoimmunology 6(6):e1259049
Kukita K et al (2015) Cancer-associated oxidase ERO1-alpha regulates the expression of MHC class I molecule via oxidative folding. J Immunol 194(10):4988–4996
Paz-Ares L et al (2018) Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med 379(21):2040–2051
Koshkin VS, Grivas P (2018) Emerging role of immunotherapy in advanced urothelial carcinoma. Curr Oncol Rep 20(6):48
Boussiotis VA (2016) Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med 375(18):1767–1778
Wolchok JD et al (2017) Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 377(14):1345–1356
Sharma P et al (2017) Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168(4):707–723
Barkal AA et al (2018) Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat Immunol 19(1):76–84
Haworth KB et al (2015) Going back to class I: MHC and immunotherapies for childhood cancer. Pediatr Blood Cancer 62(4):571–576
Chowell D et al (2018) Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359(6375):582–587
Goodman AM et al (2017) Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 16(11):2598–2608
Le DT et al (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357(6349):409–413
Yeon Yeon S et al (2019) Immune checkpoint blockade resistance-related B2M hotspot mutations in microsatellite-unstable colorectal carcinoma. Pathol Res Pract 215(1):209–214
Sade-Feldman M et al (2017) Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun 8(1):1136
Anagnostou V et al (2017) Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov 7(3):264–276
Ayers M et al (2017) IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 127(8):2930–2940
Zaretsky JM et al (2016) Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 375(9):819–829
Budczies J et al (2017) Mutation patterns in genes encoding interferon signaling and antigen presentation: a pan-cancer survey with implications for the use of immune checkpoint inhibitors. Genes Chromosomes Cancer 56(8):651–659
Ye Z et al (2018) Prevalent homozygous deletions of type I interferon and defensin genes in human cancers associate with immunotherapy resistance. Clin Cancer Res 24(14):3299–3308
Paulson KG et al (2018) Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat Commun 9(1):3868
Marijt KA, Doorduijn EM, van Hall T (2018) TEIPP antigens for T-cell based immunotherapy of immune-edited HLA class I(low) cancers. Mol Immunol. https://doi.org/10.1016/j.molimm.2018.03.029
Koyama S et al (2016) Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun 7:10501
Chojnacki S et al (2017) Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res 45(W1):W550–W553
Belmont PJ et al (2012) Regulation of microRNA expression in the heart by the ATF6 branch of the ER stress response. J Mol Cell Cardiol 52(5):1176–1182
Colangelo T et al (2016) Proteomic screening identifies calreticulin as a miR-27a direct target repressing MHC class I cell surface exposure in colorectal cancer. Cell Death Dis 7:e2120
Hisaoka M, Matsuyama A, Nakamoto M (2012) Aberrant calreticulin expression is involved in the dedifferentiation of dedifferentiated liposarcoma. Am J Pathol 180(5):2076–2083
Zhao S et al (2015) MicroRNA-148a inhibits the proliferation and promotes the paclitaxel-induced apoptosis of ovarian cancer cells by targeting PDIA3. Mol Med Rep 12(3):3923–3929
Mari L et al (2018) microRNA 125a regulates MHC-I expression on esophageal adenocarcinoma cells, associated with suppression of antitumor immune response and poor outcomes of patients. Gastroenterology 155(3):784–798
Liu Y et al (2009) Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin Med J (Engl) 122(1):10–14
Kulkarni S et al (2017) Posttranscriptional regulation of HLA-A protein expression by alternative polyadenylation signals involving the RNA-binding protein syncrip. J Immunol 199(11):3892–3899
Nachmani D et al (2014) MicroRNA editing facilitates immune elimination of HCMV infected cells. PLoS Pathog 10(2):e1003963
Kulkarni S et al (2013) Genetic interplay between HLA-C and MIR148A in HIV control and Crohn disease. Proc Natl Acad Sci USA 110(51):20705–20710
Yin P et al (2015) MiR-451 suppresses cell proliferation and metastasis in A549 lung cancer cells. Mol Biotechnol 57(1):1–11
Knox B et al (2018) A functional SNP in the 3′-UTR of TAP2 gene interacts with microRNA hsa-miR-1270 to suppress the gene expression. Environ Mol Mutagen 59(2):134–143
Acknowledgements
We would like to thank Maria Heise for excellent secretarial help in preparing the manuscript.
Funding
This work was funded by the German Research Foundation (DFG; SE 581/22-1 and RTG, 1591/2-B4), the German Israeli Foundation for Scientific Research and Development (GIF; I-37-414.11-2016), and the Mildred Scheel Foundation.
Author information
Authors and Affiliations
Contributions
Barbara Seliger planned the manuscript. All the authors contributed in writing parts of the manuscript. Michael Friedrich, Simon Jasinski-Bergner, Barbara Seliger discussed the contents, while Michael Friedrich created the figures.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This paper is a Focussed Research Review based on a presentation given at the Fourteenth Workshop & Symposium “Tumor Immunology meets Oncology” (TIMO XIV), held in Halle (Saale), Germany, 24th–26th May 2018. It is part of a series of Focussed Research Reviews and meeting report in Cancer Immunology, Immunotherapy.
Rights and permissions
About this article
Cite this article
Friedrich, M., Jasinski-Bergner, S., Lazaridou, MF. et al. Tumor-induced escape mechanisms and their association with resistance to checkpoint inhibitor therapy. Cancer Immunol Immunother 68, 1689–1700 (2019). https://doi.org/10.1007/s00262-019-02373-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-019-02373-1