Abstract
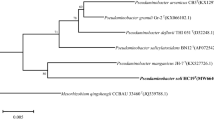
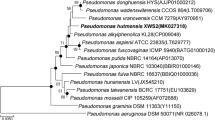
Three strains of Gram-negative bacteria designated strains H2T, H6, and H7 were isolated from bioreactors that degraded the herbicide hexazinone. Similar morphological characteristics, cellular fatty acid profiles, and 16S rRNA gene sequences show that the isolates are members of the same species. These characteristics also show that the isolates belong to the genus Pseudomonas with P. graminis, P. putida, and P. stutzeri as close relatives. The 16S rRNA gene of the H2T strain differed from that of type strains for P. graminis, P. putida, and P. stutzeri by 1.9, 2.5, and 2.7 %, respectively, indicating that the H2T, H6, and H7 strains are related to P. graminis, P. putida, and P. stutzeri but are different enough to represent a novel species. The G+C content of the three strains averaged 61.2 ± 0.8 mol% which is similar to the values reported for P. graminis (61), P. putida (61.6), and P. stutzeri (62.2–65.5). The major cellular fatty acids present in the H2T strain were C18:1 ω7c/C 18:1 ω6c (34.3 %), C16:1 ω6c/C16:1 ω7c (27.4 %), C16:0 (20.6 %), C12:0 (7.9 %), C12:0 3-OH (4.5 %), and C10:0 3-OH (3.1 %). The name Pseudomonas kuykendallii sp. nov. is proposed for these bacteria.


Similar content being viewed by others
References
Behrendt U, Ulrich A, Schumann P, Erler W, Burghardt J, Seyfarth W (1999) A taxonomic study of bacteria isolated from grasses: a proposed new species Pseudomonas graminis sp. nov. Int J Syst Bacteriol 49:297–308
Chun J, Rhee M-S, Han J-I, Bae KS (2001) Arthrobacter siderocapsulatus Dubinina and Zhdanov 1975AL is a later subjective synonym of Pseudomonas putida (Trevisan 1889) Migula 1895AL. Int J Syst Evol Microbiol 51:169–170
Buck JD (1982) Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
Gonzalez JM, Saiz-Jimenez C (2002) A fluorimetric method for the estimation of G+C mol% content in microorganisms by thermal denaturation temperature. Environ Microbiol 4:770–773
Gonzalez JM, Saiz-Jimenez C (2005) A simple fluorimetric method for the estimation of DNA–DNA relatedness between closely related microorganisms by thermal denaturation temperatures. Extremophiles 9:75–79
Greenberg AE, Clesceri LS, Eaton AD (eds) (1992) Standard methods for the examination of water and wastewater, 18th edn. American Public Health Association, Washington, pp 82–93
Hunter WJ, Follett RF, Cary JW (1997) Use of vegetable oil to stimulate denitrification and remove nitrate from flowing water. Trans ASAE 40:345–353
Hunter WJ, Manter DK (2011) Pseudomonas seleniipraecipitatus sp. nov.: a selenite reducing γ-Proteobacteria isolated from soil. Curr Microbiol 62:565–569
Hunter WJ, Shaner DL (2012) Removing hexazinone from groundwater with microbial bioreactors. Curr Microbiol 64:405–411
Jensen KIN, Kimball ER (1987) Persistence and degradation of the herbicide hexazinone in soils of lowbush blueberry fields in Nova Scotia, Canada. Bull Environ Contam Toxicol 38:232–239
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44:474–477
Lang E, Burghartz M, Spring S, Swiderski J, Spröer C (2010) Pseudomonas benzenivorans sp. nov. and Pseudomonas saponiphila sp. nov., represented by xenobiotics degrading type strains. Curr Microbiol 60:85–91
Montgomery SO, Anderson S, Waddington MG, Bardell JG, Nunn G, Foxall P (1999) Variation in bacterial interspecific distances—new rules for interpretation of 16s rDNA sequences. International Union of Microbiological Societies Meeting, Sydney, 15 Aug 1999
Palys T, Nakamura LK, Cohan FM (1997) Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol 47:1145–1156
Peterson HG, Boutin C, Freemark KE, Martin PA (1997) Toxicity of hexazinone and diquat to green algae, diatoms, cyanobacteria and duckweed. Aquat Toxicol 39:111–134
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shehata SA, Eldib MA, Abouwaly HF (1993) Effect of triazines compounds on fresh-water algae. Bull Environ Contam Toxicol 50:369–376
Spiers AJ, Buckling A, Rainey PB (2000) The causes of Pseudomonas diversity. Microbiology 146:2345–2350
Stackebrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155
Stanier RY, Palleroni NJ, Doudoroff M (1966) The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43:159–271
Stead DE (1992) Grouping of plant-pathogenic and some other Pseudomonas spp. by using cellular fatty acid profiles. Int J Syst Bacteriol 42:281–295
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Zhu Y, Li QX (2002) Movement of bromacil and hexazinone in soils of Hawaiian pineapple fields. Chemosphere 49:669–674
Acknowledgments
The authors thank Robin Montenieri and Mia Hanson for their expert technical assistance. Manufacturer and product brand names are given for the reader’s convenience and do not reflect endorsement by the US government. USDA is an equal opportunity provider and employer. This article was the work of US government employees engaged in official duties and is exempt from copyright.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hunter, W.J., Manter, D.K. Pseudomonas kuykendallii sp. nov.: A Novel γ-Proteobacteria Isolated From a Hexazinone Degrading Bioreactor. Curr Microbiol 65, 170–175 (2012). https://doi.org/10.1007/s00284-012-0141-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-012-0141-4