Abstract
Key message
Few proteomic studies have focused on the plant- Phytophthora interactions, our study provides important information regarding the use of proteomic methods for investigation of the basic mechanisms of plant- Phytophthora interactions.
Abstract
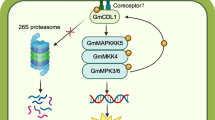
Phytophthora sojae is a fast-spreading and devastating pathogen that is responsible for root and stem rot in soybean crops worldwide. To better understand the response of soybean seedlings to the stress of infection by virulent and avirulent pathogens at the proteomic level, proteins extracted from the hypocotyls of soybean reference cultivar Williams 82 infected by P. sojae P6497 (race 2) and P7076 (race 19), respectively, were analyzed by two-dimensional gel electrophoresis. 95 protein spots were differently expressed, with 83 being successfully identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and subjected to further analysis. Based on the majority of the 83 defense-responsive proteins, and defense-related pathway genes supplemented by a quantitative reverse transcription PCR assay, a defense-related network for soybean infected by virulent and avirulent pathogens was proposed. We found reactive oxygen species (ROS) burst, the expression levels of salicylic acid (SA) signal pathway and biosynthesis of isoflavones were significantly up-regulated in the resistant soybean. Our results imply that following the P. sojae infection, ROS and SA signal pathway in soybean play the major roles in defense against P. sojae. This research will facilitate further investigation of the molecular regulatory mechanism of the defense response in soybean following infection by P. sojae.






Similar content being viewed by others
Abbreviations
- ACC:
-
1-Aminocyclopropane-1-carboxylic acid
- ACLY:
-
ATP citrate (pro-S)-lyase
- ACO:
-
1-Aminocyclopropane-1-carboxylate oxidase
- ACS:
-
1-Aminocyclopropane-1-carboxylate synthase
- AOC:
-
Allene oxide cyclase
- AOS:
-
Allene oxide synthase
- APX2:
-
Ascorbate peroxidase 2
- AsA:
-
Ascorbate
- Avr:
-
Avirulence
- CHI:
-
Chalcone isomerase
- CHR:
-
Chalcone reductase
- CHS:
-
Chalcone synthase
- citF:
-
Citrate lyase subunit alpha/citrate CoA-transferase
- COI1:
-
Coronatine insensitive 1
- CTR1:
-
Constitutive triple response 1
- DAB:
-
3,3′-diaminobenzidine
- DFR:
-
Bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase
- EDS1:
-
Enhanced disease susceptibility 1
- EDS5:
-
Enhanced disease susceptibility 5
- EIL:
-
EIN3-like
- ERF:
-
Ethylene response factor
- EIN2:
-
Ethylene insensitive 2
- EIN3:
-
Ethylene insensitive 3
- EREBP:
-
Ethylene response element binding protein
- ET:
-
Ethylene
- ETI:
-
Effector-triggered immunity
- ETR1:
-
Ethylene resistant 1
- F3H:
-
Flavanone 3-hydroxylase
- FLS:
-
Flavonol synthase
- GR:
-
Glutathione reductase
- GRP:
-
Glycine-rich protein
- GSH:
-
Glutathione
- GSSG:
-
Glutathione disulfide
- GST24:
-
Glutathione S-transferase 24
- HID:
-
2-Hydroxyisoflavanone dehydratase
- Hpa :
-
Hyaloperonospora arabidopsidis
- HR:
-
Hypersensitive response
- I2′H:
-
Isoflavone 2′-hydroxylase
- IFR:
-
Isoflavone reductase
- IFS:
-
Isoflavone synthase
- IOMT:
-
Isoflavone 7-O-methyltransferase
- JA:
-
Jasmonic acid
- JAZ:
-
Jasmonate ZIM domain-containing protein
- MAMPs or PAMPs:
-
Microbial- or pathogen-associated molecular patterns
- MnSOD:
-
Manganese superoxide dismutase
- NAC:
-
Nascent polypeptide-associated complex
- NPR1:
-
Nonexpresser of PR genes 1
- OGDH:
-
2-Oxoglutarate dehydrogenase
- OPDA:
-
Oxophytodienoic acid
- OPR3:
-
12-Oxophytodienoate reductase 3-like
- PAD4:
-
Phytoalexin deficient 4
- PAL:
-
Phenylalanine ammonia lyase
- PAPs:
-
Purple acid phosphatases
- PCD:
-
Programmed cell death
- PDF1.2:
-
Plant defensin gene
- PRRs:
-
Pattern-recognition receptors
- PTI:
-
PAMP-triggered immunity
- rab-GDI:
-
Rab GDP dissociation inhibitor
- ROS:
-
Reactive oxygen species
- Rps genes:
-
Resistance to P. sojae
- SA:
-
Salicylic acid
- SAM:
-
S-adenosyl-l-methionine
- SAMe:
-
S-adenosyl-l-methionine
- SAR:
-
Systemic acquired resistance
- SCPs:
-
Serine carboxypeptidases
- SID2:
-
SA induction-deficient 2
- UFD:
-
Ubiquitin fusion degradation
References
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Averyanov A (2009) Oxidative burst and plant disease resistance. Front Biosci (Elite Ed) 1:142–152
Baker CJ, Orlandi EW (1995) Active oxygen in plant pathogenesis. Annu Rev Phytopathol 33:299–321
Berestetskiy AO (2008) A review of fungal phytotoxins: from basic studies to practical use. Appl Biochem Microbiol 44:453–465
Bhattacharjee S, Halane MK, Kim SH, Gassmann W (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334:1405–1408
Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
De Wit PJ, Mehrabi R, Van den Burg HA, Stergiopoulos I (2009) Fungal effector proteins: past, present and future. Mol Plant Pathol 10:735–747
Dixon RA (2001) Natural products and plant disease resistance. Nature 411:843–847
Dong S, Yin W, Kong G, Yang X, Qutob D, Chen Q, Kale SD, Sui Y, Zhang Z, Dou D, Zheng X, Gijzen M, Tyler B, Wang Y (2011) Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. PLoS Pathog 7:e1002353
Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, Pauwels L, Witters E, Puga MI, Paz-Ares J, Goossens A, Reymond P, De Jaeger G, Solano R (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23:701–715
Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20:5400–5411
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815
Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, Dong X (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486:228–232
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Gordon SG, Kowitwanich K, Pipatpongpinyo W, Martin SKS, Dorrance AE (2007) Molecular marker analysis of soybean plant introductions with resistance to Phytophthora sojae. Phytopathology 97:113–118
Graham TL, Graham MY (1996) Signaling in soybean phenylpropanoid responses–dissection of primary, secondary, and conditioning effects of light, wounding, and elicitor treatments. Plant Physiol 110:1123–1133
Gutierrez-Gonzalez JJ, Wu X, Gillman JD, Lee J-D, Zhong R, Yu O, Shannon G, Ellersieck M, Nguyen HT, Sleper DA (2010) Intricate environment-modulated genetic networks control isoflavone accumulation in soybean seeds. BMC Plant Biol 10:105
Harborne JB (1990) Role of secondary metabolites in chemical defense-mechanisms in plants. Ciba F Symp 154:126–139
Horbach R, Navarro-Quesada AR, Knogge W, Deising HB (2011) When and how to kill a plant cell: infection strategies of plant pathogenic fungi. J Plant Physiol 168:51–62
Jiang RHY, Tripathy S, Govers F, Tyler BM (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci USA 105:4874–4879
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kroon LP, Brouwer H, de Cock AW, Govers F (2012) The genus phytophthora anno 2012. Phytopathology 102:348–364
Kuge S, Tachibana T, Iwai K, Naganuma A (2010) Peroxiredoxin-induced sensing and transduction of redox signal in response to oxidative stress and metabolism in yeast cells. J Pharmacol Sci 112:22
Lin F, Zhao M, Baumann DD, Ping J, Sun L, Liu Y, Zhang B, Tang Z, Hughes E, Doerge RW, Hughes TJ, Ma J (2014) Molecular response to the pathogen Phytophthora sojae among ten soybean near isogenic lines revealed by comparative transcriptomics. BMC Genom 15:18
Mano J, Belles-Boix E, Babiychuk E, Inze D, Torii Y, Hiraoka E, Takimoto K, Slooten L, Asada K, Kushnir S (2005) Protection against photooxidative injury of tobacco leaves by 2-alkenal reductase. Detoxication of lipid peroxide-derived reactive carbonyls. Plant Physiol 139:1773–1783
March-Diaz R, Garcia-Dominguez M, Lozano-Juste J, Leon J, Florencio FJ, Reyes JC (2008) Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J Cell Mol Biol 53:475–487
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498
Morris PF, Bone E, Tyler BM (1998) Chemotropic and contact responses of Phytophthora sojae hyphae to soybean isoflavonoids and artificial substrates. Plant Physiol 117:1171–1178
Mysore KS, Ryu CM (2004) Nonhost resistance: how much do we know? Trends Plant Sci 9:97–104
Nawrath C, Métraux J-P (1999) Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell Online 11:1393–1404
Neill S, Desikan R, Hancock J (2002a) Hydrogen peroxide signalling. Curr Opin Plant Biol 5:388–395
Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT (2002b) Hydrogen peroxide and nitric oxide as signalling molecules in plants. J Exp Bot 53:1237–1247
Oliva R, Win J, Raffaele S, Boutemy L, Bozkurt TO, Chaparro-Garcia A, Segretin ME, Stam R, Schornack S, Cano LM, van Damme M, Huitema E, Thines M, Banfield MJ, Kamoun S (2010) Recent developments in effector biology of filamentous plant pathogens. Cell Microbiol 12:1015
op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim CH, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, Nater M, Apel K (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15:2320–2332
Orozco-Cardenas M, Ryan CA (1999) Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci USA 96:6553–6557
Rate DN, Greenberg JT (2001) The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J 27:203–211
Rojas CM, Senthil-Kumar M, Tzin V, Mysore KS (2014) Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front Plant Sci 5:17
Schaller F, Biesgen C, Mussig C, Altmann T, Weiler EW (2000) 12-oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210:979–984
Song T, Kale SD, Arredondo FD, Shen D, Su L, Liu L, Wu Y, Wang Y, Dou D, Tyler BM (2013) Two RxLR avirulence genes in Phytophthora sojae determine soybean Rps1k-mediated disease resistance. Mol Plant Microbe Interact 26:711–720
Sugano S, Sugimoto T, Takatsuji H, Jiang CJ (2013) Induction of resistance to Phytophthora sojae in soyabean (Glycine max) by salicylic acid and ethylene. Plant Pathol 62:1048–1056
Thordal Christensen H, Zhang ZG, Wei YD, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11:1187–1194
Torres MA (2010) ROS in biotic interactions. Physiol Plant 138:414–429
Tyler BM (2007) Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol Plant Pathol 8:1–8
Tyler BM (2009) Entering and breaking: virulence effector proteins of oomycete plant pathogens. Cell Microbiol 11:13–20
van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
Vranova E, Inze D, Van Breusegem F (2002) Signal transduction during oxidative stress. J Exp Bot 53:1227–1236
Wang J, Zhang H, Allen RD (1999) Overexpression of an Arabidopsis peroxisomal ascorbate peroxidase gene in tobacco increases protection against oxidative stress. Plant Cell Physiol 40:725–732
Wang H, Waller L, Tripathy S, St Martin SK, Zhou L, Krampis K, Tucker DM, Mao Y, Hoeschele I, Saghai Maroof MA, Tyler BM, Dorrance AE (2010) Analysis of genes underlying soybean quantitative trait loci conferring partial resistance to Phytophthora sojae. Plant Genome J 3:23
Wang Q, Han C, Ferreira AO, Yu X, Ye W, Tripathy S, Kale SD, Gu B, Sheng Y, Sui Y, Wang X, Zhang Z, Cheng B, Dong S, Shan W, Zheng X, Dou D, Tyler BM, Wang Y (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23:2064–2086
Zhou L, Mideros SX, Bao L, Hanlon R, Arredondo FD, Tripathy S, Krampis K, Jerauld A, Evans C, St Martin SK, Maroof MAS, Hoeschele I, Dorrance AE, Tyler BM (2009) Infection and genotype remodel the entire soybean transcriptome. BMC Genom 10:49
Acknowledgments
This work was supported in part by grants from China National Funds for Distinguished Young Scientists (31225022) and Special Fund for Agro-Scientific Research in the Public Interest (201303018) of China to Yuanchao Wang.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. S. Judelson.
M. Jing and H. Ma contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2015_1786_MOESM1_ESM.tif
Fig. S1 The phenotypes of the infection processes during incompatible and compatible interactions between P. sojae and soybean. (TIFF 1791 kb)
299_2015_1786_MOESM2_ESM.tif
Fig. S2 Representative protein spot maps of seedling hypocotyls of soybean cultivar Williams 82 which infected by Phytophthora sojae P6497 and P7076. 2-DE was performed using 1200 µg soluble protein, nonlinear 24 cm IPG strips (pH 4-7) and 12% SDS-PAGE gels for second dimension electrophoresis. Gels were stained with CBB G-250. (TIFF 692 kb)
299_2015_1786_MOESM3_ESM.tif
Fig. S3 Identified proteins involved in the metabolic pathway network. SOD, superoxide dismutase; GSSG, glutathione disulfide; GSH, glutathione; ACS, 1-aminocyclopropane-1-carboxylate synthase; OGDH, 2-oxoglutarate dehydrogenase; ACLY, ATP citrate (pro-S)-lyase; and citF, citrate lyase subunit alpha / citrate CoA-transferase. Red italics indicate the protein spots identified in this study. (TIFF 422 kb)
299_2015_1786_MOESM4_ESM.tif
Fig. S4 Histochemical identification of H2O2 by DAB staining in soybean hypocotyls. A, C: soybean hypocotyls at 12 and 24 h after inoculation with zoospores were stained by DAB; the incompatible interaction; B, D: soybean hypocotyls at 12 and 24 h after inoculation with zoospores were stained by DAB; the compatible interaction. Bar, 50 μm. (TIFF 5929 kb)
299_2015_1786_MOESM5_ESM.tif
Fig. S5 The protein–protein interaction in seedling hypocotyls from the soybean cultivar Williams 82 infected by P. sojae P6497 and P7076. Analysis of a predicted protein–protein interaction network using STRING 9.1 (http://string-db.org). Arabidopsis thaliana and a confidence level of 0.4 were used as analysis parameters. Different-colored lines represent the types of evidence used to predict the associations: gene fusion (red), neighborhood (green), co-occurrence across genomes (blue), co-expression (black), experimental (purple), association in curated databases (light blue) or co-mentioned in PubMed abstracts (yellow). Five clusters of highly interacting protein nodes are marked with circles and include the proteins involved in amino acid and nitrogen metabolism, signal transduction, redox homeostasis, carbohydrate metabolism, and secondary metabolism. (TIFF 2125 kb)
299_2015_1786_MOESM6_ESM.docx
Table S1 The descriptions of the 83 differentially expressed proteins identified by MS/MS in the seedling hypocotyls from soybean cultivar Williams 82. (DOCX 47 kb)
299_2015_1786_MOESM8_ESM.rar
Supplementary material 2 All 2-DE maps of seedling hypocotyls of soybean cultivar Williams 82, which infected by Phytophthora sojae P6497 and P7076. (RAR 17807 kb)
Rights and permissions
About this article
Cite this article
Jing, M., Ma, H., Li, H. et al. Differential regulation of defense-related proteins in soybean during compatible and incompatible interactions between Phytophthora sojae and soybean by comparative proteomic analysis. Plant Cell Rep 34, 1263–1280 (2015). https://doi.org/10.1007/s00299-015-1786-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1786-9