Abstract
Objectives
To compare diagnostic performance for pulmonary invasive adenocarcinoma among radiologists with and without three-dimensional convolutional neural network (3D-CNN).
Methods
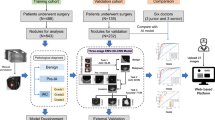
Enrolled were 285 patients with adenocarcinoma in situ (AIS, n = 75), minimally invasive adenocarcinoma (MIA, n = 58), and invasive adenocarcinoma (IVA, n = 152). A 3D-CNN model was constructed with seven convolution-pooling and two max-pooling layers and fully connected layers, in which batch normalization, residual connection, and global average pooling were used. Only the flipping process was performed for augmentation. The output layer comprised two nodes for two conditions (AIS/MIA and IVA) according to prognosis. Diagnostic performance of the 3D-CNN model in 285 patients was calculated using nested 10-fold cross-validation. In 90 of 285 patients, results from each radiologist (R1, R2, and R3; with 9, 14, and 26 years of experience, respectively) with and without the 3D-CNN model were statistically compared.
Results
Without the 3D-CNN model, accuracy, sensitivity, and specificity of the radiologists were as follows: R1, 70.0%, 52.1%, and 90.5%; R2, 72.2%, 75%, and 69%; and R3, 74.4%, 89.6%, and 57.1%, respectively. With the 3D-CNN model, accuracy, sensitivity, and specificity of the radiologists were as follows: R1, 72.2%, 77.1%, and 66.7%; R2, 74.4%, 85.4%, and 61.9%; and R3, 74.4%, 93.8%, and 52.4%, respectively. Diagnostic performance of each radiologist with and without the 3D-CNN model had no significant difference (p > 0.88), but the accuracy of R1 and R2 was significantly higher with than without the 3D-CNN model (p < 0.01).
Conclusions
The 3D-CNN model can support a less-experienced radiologist to improve diagnostic accuracy for pulmonary invasive adenocarcinoma without deteriorating any diagnostic performances.
Key Points
• The 3D-CNN model is a non-invasive method for predicting pulmonary invasive adenocarcinoma in CT images with high sensitivity.
• Diagnostic accuracy by a less-experienced radiologist was better with the 3D-CNN model than without the model.






Similar content being viewed by others
Abbreviations
- 3D:
-
Three-dimensional
- AIS:
-
Adenocarcinoma in situ
- AUC:
-
Area under the curve
- CI:
-
Confidence interval
- CNN:
-
Convolutional neural network
- GGN:
-
Ground-glass nodule
- IVA:
-
Invasive adenocarcinoma
- MIA:
-
Minimally invasive adenocarcinoma
- WHO:
-
World Health Organization
References
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69:7–34
Goldstraw P, Chansky K, Crowley J et al (2016) The IASLC Lung Cancer Staging Project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol 11:39–51
Travis WD, Asamura H, Bankier AA et al (2016) The IASLC Lung Cancer Staging Project: proposals for coding T categories for subsolid nodules and assessment of tumor size in part-solid tumors in the forthcoming eighth edition of the TNM classification of lung cancer. J Thorac Oncol 11:1204–1223
Borczuk AC, Qian F, Kazeros A et al (2009) Invasive size is an independent predictor of survival in pulmonary adenocarcinoma. Am J Surg Pathol 33:462–469
Wilshire CL, Louie BE, Manning KA et al (2015) Radiologic evaluation of small lepidic adenocarcinomas to guide decision making in surgical resection. Ann Thorac Surg 100:979–988
Colby TV, Noguchi M, Henschke H et al (2004) Adenocarcinoma. In: Travis WD, Brambilla HK, Muller-Hermelink K, Harris CC (eds) World Health Organization classification of tumours. Tumors of the lung, pleura, thymus and heart, 1st ed. IARC, Lyon, pp 35–44
Naidich DP, Bankier AA, MacMahon H et al (2013) Recommendations for the management of subsolid pulmonary nodules detected at CT: a statement from the Fleischner Society. Radiology 266:304–317
Maeshima AM, Tochigi N, Yoshida A et al (2010) Histological scoring for small lung adenocarcinomas 2cm or less in diameter: a reliable prognostic indicator. J Thorac Oncol 5:333–339
Tsutani Y, Miyata Y, Mimae T et al (2013) The prognostic role of pathologic invasive component size, excluding lepidic growth, in stage I lung adenocarcinoma. J Thorac Cardiovasc Surg 146:580–585
Niioka H, Asatani S, Yoshimura A, Ohigashi H, Tagawa S, Miyake J (2018) Classification of C2C12 cells at differentiation by convolutional neural network of deep learning using phase contrast images. Hum Cell 31:87–93
Ehteshami Bejnordi B, Veta M, Johannes van Diest P et al (2017) Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318:2199–2210
Gulshan V, Peng L, Coram M et al (2016) Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316:2402–2410
Russakovsky O, Deng J, Su H et al (2015) ImageNet large scale visual recognition challenge. Int J Comput Vis 115:211–252
Esteva A, Kuprel B, Novoa RA et al (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature. 542:115–118
Kermany DS, Goldbaum M, Cai W et al (2018) Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 172:1122–1131
Hwang EJ, Hong JH, Lee KH et al (2020) Deep learning algorithm for surveillance of pneumothorax after lung biopsy: a multicenter diagnostic cohort study. Eur Radiol 30:3660–3671
Gong L, Jiang S, Yang Z, Zhang G, Wang L (2019) Automated pulmonary nodule detection in CT images using 3D deep squeeze-and-excitation networks. Int J Comput Assist Radiol Surg 14:1969–1979
Xie Y, Xia Y, Zhang J et al (2019) Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans Med Imaging 38:991–1004
Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, Kluger Y (2018) DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol 18:24
She Y, Jin Z, Wu J et al (2020) Development and validation of a deep learning model for non–small cell lung cancer survival. JAMA Netw Open 3:e205842
Travis WD, Brambilla E, Nicholson AG et al (2015) The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 10:1243–1260
Borczuk AC (2016) Prognostic considerations of the new World Health Organization classification of lung adenocarcinoma. Eur Respir Rev 25:364–371
Yanagawa M, Niioka H, Hata A et al (2019) Application of deep learning (3-dimensional convolutional neural network) for the prediction of pathological invasiveness in lung adenocarcinoma: a preliminary study. Medicine (Baltimore) 98(25):e16119
Maclure M, Willett WC (1987) Misinterpretation and misuse of the kappa statistic. Am J Epidemiol 126:161–169
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 44:837–845
Jin H, Li Z, Tong R, Lin L (2018) A deep 3D residual CNN for false-positive reduction in pulmonary nodule detection. Med Phys 45:2097–2107
Gong J, Liu J, Hao W et al (2020) A deep residual learning network for predicting lung adenocarcinoma manifesting as ground-glass nodule on CT images. Eur Radiol 30(4):1847–1855
Aberle DR, Adams AM, Berg CD et al (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 365:395–409
McKee BJ, Regis SM, McKee AB, Flacke S, Wald C (2015) Performance of ACR Lung-RADS in a clinical CT lung screening program. J Am Coll Radiol 12:273–276
van Riel SJ, Jacobs C, Scholten ET et al (2019) Observer variability for Lung-RADS categorisation of lung cancer screening CTs: impact on patient management. Eur Radiol 29:924–931
Ardila D, Kiraly AP, Bharadwaj S et al (2019) End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 25:954–961
Funding
This work was supported by a Grant-in-Aid for Scientific Research (C) (Grant Number JP18K07713). Masahiro Yanagawa, Hirohiko Niioka, Noriyuki Tomiyama, and Jun Miyake received a Grant-in-Aid for Scientific Research (C).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Noriyuki Tomiyama.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the institutional review board.
Ethical approval
Institutional review board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary materials
Supplementary Fig. 1
How to separate data in nested 10-fold cross validation. Predictions from nine models were assigned to the same test data. The average of the predictions from the nine models was calculated and a prediction label was given to each test datum (DOCX 461 kb)
Rights and permissions
About this article
Cite this article
Yanagawa, M., Niioka, H., Kusumoto, M. et al. Diagnostic performance for pulmonary adenocarcinoma on CT: comparison of radiologists with and without three-dimensional convolutional neural network. Eur Radiol 31, 1978–1986 (2021). https://doi.org/10.1007/s00330-020-07339-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07339-x