Abstract
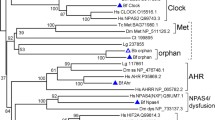
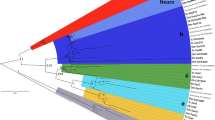
Genes of the iroquois (Iro/Irx) family are highly conserved from Drosophila to mammals and they have been implicated in a number of developmental processes. In flies, the Iro genes participate in patterning events in the early larva and in imaginal disk specification. In vertebrates, the Irx genes regulate developmental events during gastrulation, nervous system regionalization, activation of proneural genes and organ patterning. The Iro genes in Drosophila and the Irx genes of mammals show a clustered organization in the genome. Flies have a single cluster comprising three genes while mammals have two clusters also having three genes each. Moreover, experimental evidence in flies shows that transcriptional regulatory elements are shared among genes within the Iro cluster, suggesting that the same may be true in vertebrates. To date, the genomic organization of the Irx genes in non-mammalian species has not been studied. In this work, we have isolated the irx5b gene from zebrafish, Danio rerio, and have characterized its expression pattern. Furthermore, we have identified the complete set of Irx genes in two fish species, the zebrafish and pufferfish, Takifugu rubripes, and have determined the genomic organization of these genes. Our analysis indicates that early in fish evolutionary history, the Irx gene clusters have been duplicated and that subsequent events have maintained the clustered organization for some of the genes, while others have been lost. In total there are 11 existing Irx genes in zebrafish and 10 in pufferfish. We propose a new nomenclature for the zebrafish Irx genes based on the analysis of their sequences and their genomic relationships.




Similar content being viewed by others
References
Adachi J, Hasegawa M (1996) MOLPHY version 2.3: programs for molecular phylogenetics based on maximum likelihood. Comput Sci Monogr Inst Stat Math 28:1–50
Aparicio S (2000) Vertebrate evolution: recent perspectives from fish. Trends Genet 16:54–56
Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL (1999) Regulation of chamber-specific gene expression in the developing heart by Irx4. Science 283:1161–1164
Bellefroid EJ, Kobbe A, Gruss P, Pieler T, Gurdon JB, Papalopulu N (1998) Xiro3 encodes a Xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO J 17:191–203
Bosse A, Stoykova A, Nieselt-Struwe K, Chowdhury K, Copeland N, Jenkins NA, Gruss P (2000) Identification of a novel mouse Iroquois homeobox gene, Irx5, chromosomal localization of all members of the mouse Iroquois gene family. Dev Dyn 218:160–174
Briscoe J, Pierani A, Jessell TM, Ericson J (2000) A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101:435–445
Burglin TR (1997) Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res 25:4173–4180
Cavodeassi F, Diez del Corral R, Campuzano S, Dominguez M (1999) Compartments and organizing boundaries in the Drosophila eye: the role of the homeodomain Iroquois proteins. Development 126:4833–4942
Cavodeassi F, Modolell J, Gómez-Skarmeta JL (2001) The Iroquois family of genes: from body building to neural patterning. Development 128:2847–2855
Dambly-Chaudière C, Leyns L (1992) The determination of sense organs in Drosophila: a search for interacting genes. Int J Dev Biol 36:85–91
Diez del Corral R, Aroca P, Gómez-Skarmeta JL, Cadoveassi F, Modolell J (1999) The iroquois homeodomain proteins are required to specify body wall identify in Drosophila. Genes Dev 13:1754–1761
Dildrop R, Rüther U (2004) Organization of iroquois genes in fish. Dev Genes Evol DOI 10.1007/s00427-004-0402-8
Duboule D (1998) Vertebrate Hox gene regulation: clustering and/or colinearity? Curr Opin Genet Dev 8:514–518
Felsenstein J (1993) PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle
Felsenstein J (1996) Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol 266:418–427
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545
Funayama N, Sato Y, Matsumoto K, Ogura T, Takahashi Y (1999) Coelom formation: binary decision of the lateral plate mesoderm is controlled by the ectoderm. Development 126:4129–4138
Garriock RJ, Vokes SA, Small EM, Larson R, Krieg PA (2001) Developmental expression of the Xenopus Iroquois-family homeobox genes, Irx4 and Irx5. Dev Genes Evol 211:257–260
Gómez-Skarmeta JL, Modolell J (2002) Iroquois genes: genomic organization and function in vertebrate neural development. Curr Opin Genet Dev 12:403–408
Gómez-Skarmeta JL, Diez del Corral R, de la Calle-Mustienes E, Ferrés-Marcó D, Modolell J (1996) araucan and caupolican, two members of the novel Iroquois complex, encode homeoproteins that control proneural and vein forming genes. Cell 85:95–105
Gómez-Skarmeta JL, Glavic A, de la Calle-Mustienes E, Modollel J, Mayor R (1998) Xiro, a Xenopus homolog of the Drosophila Iroquois complex genes, controls development al the neural plate. EMBO J 17:181–190
Gómez-Skarmeta JL, de la Calle-Mustienes E, Modollel J (2001) The Wnt-activated Xiro1 gene encodes a repressor that is essential for neural development and down regulates BMP4. Development 128:551–560
Goriely A, Diez del Corral R, Storey KG (1999) c-Irx2 expression reveals an early subdivision of the neural plate in the chick embryo. Mech Dev 87:203–206
Houweling AC, Dildrop R, Peters T, Mummenhoff J, Moorman AFM, Rüther U, Christoffels VM (2001) Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech Dev 107:169–174
Itoh M, Kudoh T, Dedekian M, Kim CH, Chitnis AB (2002) A role for iro1 and iro7 in the establishment of an anteroposterior compartment of the ectoderm adjacent to the midbrain-hindbrain boundary. Development 129:2317–2327
Jowett T, Lettice L (1994) Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin-and fluorescein-labeled probes. Trends Genet 10:73–74
Kobayashi D, Kobayashi M, Matsumoto K, Ogura T, Nakafuku M, Shimamura K (2002) Early subdivisions in the neural plate define distinct competence for inductive signals. Development 129:83–93
Kudoh T, Dawid IB (2001) Role of the Iroquois3 homeobox gene in organizer formation. Proc Natl Acad Sci USA 98:7852–7857
Kwok C, Korn R, Davis M, Burt D, Paw B, Zon L, Goodfellow P, Schmitt K (1998) Characterization of whole genome radiation hybrid mapping resources for non-mammalian vertebrates. Nucleic Acids Res 26:3562–3566
Lecaudey V, Thisse C, Thisse B, Schneider-Maunoury S (2001) Sequence and expression pattern of ziro7, a novel, divergent zebrafish iroquois homeobox gene. Mech Dev 109:383–388
Leyns L, Gómez-Skarmeta JL, Dambly-Chaudière C (1996) iroquois: a prepattern gene that controls the formation of bristles on the thorax of Drosophila. Mech Dev 59:63–72
McNeil H, Yang CH, Brosky M, Ungos J, Simon MA (1997) mirror encodes a novel PBX-class homeoprotein that functions in the definition of dorso-ventral border of Drosophila eye. Genes Dev 11:1073–1082
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Peters T, Dildrop R, Ausmeier K, Ruther U (2000) Organization of the mouse Iroquois homeobox genes in two clusters suggest a conserved regulation and function in vertebrate development. Genome Res 10:1453–1462
Prince VE, Pickett FB (2002) Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet 3:827–837
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Tan JT, Korzh V, Gong Z (1999) Expression of a zebrafish iroquois homeobox gene, Ziro3, in the midline axial structures and central nervous system. Mech Dev 87:165–168
Taylor JS, Van de Peer Y, Braasch I, Meyer A (2001) Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans R Soc Lond B Biol Sci 356:1661–1679
Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y (2003) Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res 13:382–390
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by its quality analysis tools. Nucleic Acids Res 25:4876–4882
Wang X, Emelyanov A, Sleptsova-Friedrich I, Korzh V, Gong Z (2001) Expression of two novel zebrafish Iroquois homologues (ziro1 and ziro5) during early development of axial structures and central nervous system. Mech Dev 105:191–195
Westerfield M (1995) The zebrafish book. University of Oregon Press, Eugene, Ore.
Acknowledgements
We thank Renate Dildrop and Ulrich Rüther for providing a copy of their manuscript prior to publication. X. Wang kindly provided probes. We are also grateful to Peng Jinrong for clone 012-H09-2. We wish to thank Gabino Sánchez-Pérez for help with phylogenetic analysis. Carolina Achondo and Florencio Espinoza provided technical help. C.G.F. and M.A. were supported by grants from Fondecyt (1031003), the ICM (P02-050) and by a grant from the UNAB (DI 10-03). M.M. and J.L.G.-S. were supported by grants from the Spanish Ministry of Science and Technology (BMC2002-03558 and BMC2001-2122, respectively). In addition, M.A. and J.L.G.-S. were supported by a CONICYT-CSIC collaboration grant (2003CL00021).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by D. Tautz
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Feijóo, C.G., Manzanares, M., de la Calle-Mustienes, E. et al. The Irx gene family in zebrafish: genomic structure, evolution and initial characterization of irx5b . Dev Genes Evol 214, 277–284 (2004). https://doi.org/10.1007/s00427-004-0401-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-004-0401-9