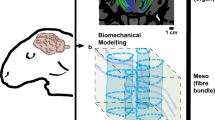
Abstract
Understanding the intricate three-dimensional relationship between fiber bundles and subcortical nuclei is not a simple task. It is of paramount importance in neurosciences, especially in the field of functional neurosurgery. The current methods for in vivo and post mortem fiber tract visualization have shortcomings and contributions to the field are welcome. Several tracts were chosen to implement a new technique to help visualization of white matter tracts, using high-thickness histology and dark field images. Our study describes the use of computational fluid dynamic simulations for visualization of 3D fiber tracts segmented from dark field microscopy in high-thickness histological slices (histological mesh tractography). A post mortem human brain was MRI scanned prior to skull extraction, histologically processed and serially cut at 430 µm thickness as previously described by our group. High-resolution dark field images were used to segment the outlines of the structures. These outlines served as basis for the construction of a 3D structured mesh, were a Finite Volume Method (FVM) simulation of water flow was performed to generate streamlines representing the geometry. The simulations were accomplished by an open source computer fluid dynamics software. The resulting simulation rendered a realistic 3D impression of the segmented anterior commissure, the left anterior limb of the internal capsule, the left uncinate fascicle, and the dentato-rubral tracts. The results are in line with clinical findings, diffusion MR imaging and anatomical dissection methods.





Similar content being viewed by others
References
Ahrens J, Geveci B, Law C (2005) Paraview: an end-user tool for large-data visualization. Visualization handbook. Elsevier, Amsterdam, pp 717–731
Alegro M, Loring B, Alho E, et al (2016) Multimodal whole brain registration: mri and high resolution histology. http://www.cv-foundation.org/openaccess/content_cvpr_2016_workshops/w15/papers/Alegro_Multimodal_Whole_Brain_CVPR_2016_paper.pdf. Accessed 9 Sep 2016
Alexander AL, Hasan KM, Lazar M et al (2001) Analysis of partial volume effects in diffusion-tensor MRI. Magn Reson Med 45:770–780. https://doi.org/10.1002/mrm.1105
Alho ATDL, Hamani C, Alho EJL et al (2017a) Magnetic resonance diffusion tensor imaging for the pedunculopontine nucleus: proof of concept and histological correlation. Brain Struct Funct. https://doi.org/10.1007/s00429-016-1356-0
Alho EJL, Alho ATDL, Grinberg L et al (2017b) High thickness histological sections as alternative to study the three-dimensional microscopic human sub-cortical neuroanatomy. Brain Struct Funct. https://doi.org/10.1007/s00429-017-1548-2
Andral G (1837) Die Krankheiten des Gehirns I. die Krankheiten der Gehirnhäute, die Hyperämien und Hämorrhagien des Gehirns enthaltend
Avants BB, Epstein CL, Grossman M, Gee JC (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12:26–41. https://doi.org/10.1016/j.media.2007.06.004
Axer H, Berks G, Keyserlingk DGV (2000) Visualization of nerve fiber orientation in gross histological sections of the human brain. Microsc Res Tech 51:481–492. https://doi.org/10.1002/1097-0029(20001201)51:5%3c481::AID-JEMT11%3e3.0.CO;2-N
Basser PJ, Özarslan E (2010) Anisotropic diffusion: from the apparent diffusion coefficient to the apparent diffusion tensor. In: diffusion MRI theory, methods and applications. Oxford: Oxford University Press
Bergström RM (1969) An entropy model of the developing brain. Dev Psychobiol 2:139–152. https://doi.org/10.1002/dev.420020304
Bowden DM, Martin RF (1995) Neuronames brain hierarchy. NeuroImage 2:63–83. https://doi.org/10.1006/nimg.1995.1009
Calabrese E (2016) Diffusion tractography in deep brain stimulation surgery: a review. Front Neuroanat. https://doi.org/10.3389/fnana.2016.00045
Catani M, Howard RJ, Pajevic S, Jones DK (2002) Virtual in vivo interactive dissection of white matter fasciculi in the human brain. NeuroImage 17:77–94
Chung K, Wallace J, Kim S-Y et al (2013) Structural and molecular interrogation of intact biological systems. Nature 497:332–337. https://doi.org/10.1038/nature12107
Coenen VA, Schlaepfer TE, Reinacher PC et al (2019) Machine learning-aided personalized DTI tractographic planning for deep brain stimulation of the superolateral medial forebrain bundle using HAMLET. Acta Neurochir (Wien) 161:1559–1569. https://doi.org/10.1007/s00701-019-03947-9
Coenen VA, Schlaepfer TE, Sajonz B et al (2020) Tractographic description of major subcortical projection pathways passing the anterior limb of the internal capsule. Corticopetal organization of networks relevant for psychiatric disorders. NeuroImage Clin 25:102165. https://doi.org/10.1016/j.nicl.2020.102165
Coenen VA, Varkuti B, Parpaley Y et al (2017) Postoperative neuroimaging analysis of DRT deep brain stimulation revision surgery for complicated essential tremor. Acta Neurochir (Wien) 159:779–787. https://doi.org/10.1007/s00701-017-3134-z
Cui SZ, Li EZ, Zang YF et al (2000) Both sides of human cerebellum involved in preparation and execution of sequential movements. NeuroReport 11:3849–3853. https://doi.org/10.1097/00001756-200011270-00049
Dejerine J (1849–1917) A du texte (1895) Anatomie des centres nerveux. Anatomie du rhombencéphale / par J. Dejerine,... ; avec la collaboration de Mme Dejerine-Klumpke,...
Dos Santos Ghilardi MG, Ibarra M, Alho EJL et al (2018) Double-target DBS for essential tremor: 8-contact lead for cZI and Vim aligned in the same trajectory. Neurology 90:476–478. https://doi.org/10.1212/WNL.0000000000005076
Forel A (1877) Untersuchungen über die Haubenregion und ihre oberen verknüpfungen im Gehirne des Menschen und einiger Säugethiere: mit Beiträgen zu den Methoden der Gehirnuntersuchung ... G. Bernstein
Fortier PA, Kalaska JF, Smith AM (1989) Cerebellar neuronal activity related to whole-arm reaching movements in the monkey. J Neurophysiol 62:198–211. https://doi.org/10.1152/jn.1989.62.1.198
Fraioli B, Guidetti B (1975) Effects of stereotactic lesions of the dentate nucleus of the cerebellum in man. Appl Neurophysiol 38:81–90
Friedman DI, Johnson JK, Chorsky RL, Stopa EG (1991) Labeling of human retinohypothalamic tract with the carbocyanine dye, DiI. Brain Res 560:297–302
Gray H, Lewis WH (1918) Anatomy of the human body. Lea & Febiger, Philadelphia
Horn A, Li N, Dembek TA et al (2019) Lead-DBS v2: Towards a comprehensive pipeline for deep brain stimulation imaging. NeuroImage 184:293–316. https://doi.org/10.1016/j.neuroimage.2018.08.068
Immisch I, Quintern J, Straube A (2003) Unilateral cerebellar lesions influence arm movements bilaterally. NeuroReport 14:837–840. https://doi.org/10.1097/00001756-200305060-00012
Kamali A, Kramer LA, Frye RE et al (2010) Diffusion tensor tractography of the human brain cortico-ponto-cerebellar pathways: a quantitative preliminary study. J Magn Reson Imaging JMRI 32:809–817. https://doi.org/10.1002/jmri.22330
Kawashima R, Matsumura M, Sadato N et al (1998) Regional cerebral blood flow changes in human brain related to ipsilateral and contralateral complex hand movements–a PET study. Eur J Neurosci 10:2254–2260. https://doi.org/10.1046/j.1460-9568.1998.00237.x
Keser Z, Hasan KM, Mwangi BI et al (2015) Diffusion tensor imaging of the human cerebellar pathways and their interplay with cerebral macrostructure. Front Neuroanat. https://doi.org/10.3389/fnana.2015.00041
Khan AR, Cornea A, Leigland LA et al (2015) 3D structure tensor analysis of light microscopy data for validating diffusion MRI. NeuroImage 111:192–203. https://doi.org/10.1016/j.neuroimage.2015.01.061
Kinoshita H, Oku N, Hashikawa K, Nishimura T (2000) Functional brain areas used for the lifting of objects using a precision grip: a PET study. Brain Res 857:119–130. https://doi.org/10.1016/s0006-8993(99)02416-6
Klingler J, Klingler JP (1935) Erleichterung der makroskopischen Praeparation des Gehirns durch den Gefrierprozess
Kwon HG, Hong JH, Hong CP et al (2011) Dentatorubrothalamic tract in human brain: diffusion tensor tractography study. Neuroradiology 53:787–791. https://doi.org/10.1007/s00234-011-0878-7
Mai JK, Paxinos G, Voss T (2008) Atlas of the human brain. Academic Press, New York, London
Meola A, Comert A, Yeh F-C et al (2016) The nondecussating pathway of the dentatorubrothalamic tract in humans: human connectome-based tractographic study and microdissection validation. J Neurosurg 124:1406–1412. https://doi.org/10.3171/2015.4.JNS142741
Mollink J, van Baarsen KM, Dederen PJWC et al (2016) Dentatorubrothalamic tract localization with postmortem MR diffusion tractography compared to histological 3D reconstruction. Brain Struct Funct 221:3487–3501. https://doi.org/10.1007/s00429-015-1115-7
Mori S, Crain BJ, Chacko VP, van Zijl PC (1999) Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol 45:265–269
Mori S, Zhang J (2006) Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 51:527–539. https://doi.org/10.1016/j.neuron.2006.08.012
Nuttin B, Cosyns P, Demeulemeester H et al (1999) Electrical stimulation in anterior limbs of internal capsules in patients with obsessive-compulsive disorder. Lancet Lond Engl 354:1526. https://doi.org/10.1016/S0140-6736(99)02376-4
Nuttin BJ, Gabriëls LA, Cosyns PR et al (2003) Long-term electrical capsular stimulation in patients with obsessive-compulsive disorder. Neurosurgery 52:1263–1274. https://doi.org/10.1227/01.NEU.0000064565.49299.9A
Oh SW, Harris JA, Ng L et al (2014) A mesoscale connectome of the mouse brain. Nature 508:207. https://doi.org/10.1038/nature13186
Parent M, Parent A (2004) The pallidofugal motor fiber system in primates. Parkinsonism Relat Disord 10:203–211. https://doi.org/10.1016/j.parkreldis.2004.02.007
Patenaude B, Smith SM, Kennedy DN, Jenkinson M (2011) A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage 56:907–922. https://doi.org/10.1016/j.neuroimage.2011.02.046
Pierpaoli C, Jezzard P, Basser PJ et al (1996) Diffusion tensor MR imaging of the human brain. Radiology 201:637–648. https://doi.org/10.1148/radiology.201.3.8939209
Riley H (1943) An atlas of the basal ganglia, brain stem and spinal cord, based on myelin-stained material. Arch Neurol Psychiatry 50:741–741. https://doi.org/10.1001/archneurpsyc.1943.02290240125015
Rojas-Vite G, Coronado-Leija R, Narvaez-Delgado O et al (2019) Histological validation of per-bundle water diffusion metrics within a region of fiber crossing following axonal degeneration. NeuroImage 201:116013. https://doi.org/10.1016/j.neuroimage.2019.116013
Safadi Z, Grisot G, Jbabdi S et al (2018) Functional segmentation of the anterior limb of the internal capsule: linking white matter abnormalities to specific connections. J Neurosci 38:2106–2117. https://doi.org/10.1523/JNEUROSCI.2335-17.2017
Saliani A, Perraud B, Duval T et al (2017) Axon and myelin morphology in animal and human spinal cord. Front Neuroanat 11:129. https://doi.org/10.3389/fnana.2017.00129
Schaltenbrand G, Hassler R, Wahren W (1977) Atlas for stereotaxy of the human brain: with an accompanying guide. Thieme, Stuttgart
Schilling KG, By S, Feiler HR et al (2019) Diffusion MRI microstructural models in the cervical spinal cord—Application, normative values, and correlations with histological analysis. NeuroImage 201:116026. https://doi.org/10.1016/j.neuroimage.2019.116026
Schulz G, Crooijmans HJA, Germann M et al (2011) Three-dimensional strain fields in human brain resulting from formalin fixation. J Neurosci Methods 202:17–27. https://doi.org/10.1016/j.jneumeth.2011.08.031
Silva SM, Andrade JP (2016) Neuroanatomy: the added value of the Klingler method. Ann Anat Anat Anz 208:187–193. https://doi.org/10.1016/j.aanat.2016.06.002
Simmons DM, Swanson LW (2009) Comparing histological data from different brains: sources of error and strategies for minimizing them. Brain Res Rev 60:349–367. https://doi.org/10.1016/j.brainresrev.2009.02.002
Sinke MRT, Otte WM, Christiaens D et al (2018) Diffusion MRI-based cortical connectome reconstruction: dependency on tractography procedures and neuroanatomical characteristics. Brain Struct Funct 223:2269–2285. https://doi.org/10.1007/s00429-018-1628-y
Soteropoulos DS, Baker SN (2008) Bilateral representation in the deep cerebellar nuclei. J Physiol 586:1117–1136. https://doi.org/10.1113/jphysiol.2007.144220
Visser E, Keuken MC, Douaud G et al (2016) Automatic segmentation of the striatum and globus pallidus using MIST: multimodal image segmentation tool. NeuroImage 125:479–497. https://doi.org/10.1016/j.neuroimage.2015.10.013
Vogt O (1904) Neurobiologische Arbeiten: ser. Beitra ge zur Hirnfaserlehre. Fischer
Von Der Heide RJ, Skipper LM, Klobusicky E, Olson IR (2013) Dissecting the uncinate fasciculus: disorders, controversies and a hypothesis. Brain 136:1692–1707. https://doi.org/10.1093/brain/awt094
Wakely J (1991) A colour atlas of the brain and spinal cord, 1st edn. Mosby, London
Wang H, Fu Y, Zickmund P et al (2005) Coherent anti-stokes Raman scattering imaging of axonal myelin in live spinal tissues. Biophys J 89:581–591. https://doi.org/10.1529/biophysj.105.061911
Wiegell MR, Larsson HB, Wedeen VJ (2000) Fiber crossing in human brain depicted with diffusion tensor MR imaging. Radiology 217:897–903. https://doi.org/10.1148/radiology.217.3.r00nv43897
Yeh F-C, Panesar S, Fernandes D et al (2018) Population-averaged atlas of the macroscale human structural connectome and its network topology. NeuroImage 178:57–68. https://doi.org/10.1016/j.neuroimage.2018.05.027
Yeh F-C, Tseng W-YI (2011) NTU-90: a high angular resolution brain atlas constructed by q-space diffeomorphic reconstruction. NeuroImage 58:91–99. https://doi.org/10.1016/j.neuroimage.2011.06.021
Yeh F-C, Wedeen VJ, Tseng W-YI (2010) Generalized q-sampling imaging. IEEE Trans Med Imaging 29:1626–1635. https://doi.org/10.1109/TMI.2010.2045126
Acknowledgments
The authors would like to thank the team participating on the São Paulo-Würzburg collaborative project. This includes all members of the Brain Bank of the Brazilian Aging Brain Research Group (BBBABSG) of the University of São Paulo Medical School, Mrs. E. Broschk and Mrs. A. Bahrke from the Morphological Brain Research Unit of the University of Würzburg, Germany.
Funding
This study was supported by resources from the University of Sao Paulo School of Medicine, Brazil and University of Würzburg, Germany. The author Eduardo Joaquim Lopes Alho was supported by a scholarship from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) agency, Brazil, for doctoral studies at the University of Würzburg, Germany. The authors do not have personal financial or institutional interest in any of the drugs, materials, or devices described in this article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors disclose any actual or potential conflict of interest including any financial, personal or other relationships with other people or organizations within three years of beginning the submitted work that could inappropriately influence, or be perceived to influence, their work.
Human rights
The work described has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Alho, E.J.L., Fonoff, E.T., Di Lorenzo Alho, A.T. et al. Use of computational fluid dynamics for 3D fiber tract visualization on human high-thickness histological slices: histological mesh tractography. Brain Struct Funct 226, 323–333 (2021). https://doi.org/10.1007/s00429-020-02187-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-020-02187-3