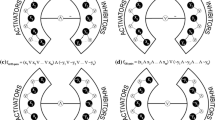
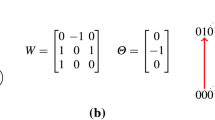
Abstract
Biological regulatory network can be modeled through a set of Boolean functions. These set of functions enable graph representation of the network structure, and hence, the dynamics of the network can be seen easily. In this article, the regulations of such network have been explored in terms of interaction graph. With the help of Boolean function decomposition, this work presents an approach for construction of interaction graphs. This decomposition technique is also used to reduce the network state space of the cell cycle network of fission yeast for finding the singleton attractors. Some special classes of Boolean functions with respect to the interaction graphs have been discussed. A unique recursive procedure is devised which uses the Cartesian product of sets starting from the set of one-variable Boolean function. Interaction graphs generated with these Boolean functions have only positive/negative edges, and the corresponding state spaces have periodic attractors with length one/two.












Similar content being viewed by others
References
Akutsu T, Kuhara S, Maruyama O, Miyano S (1998) A system for identifying genetic networks from gene expression patterns produced by gene disruptions and overexpressions. Genome Inf 9:151–160
Ay F, Xu F, Kahveci T (2009) Scalable steady state analysis of boolean biological regulatory networks. PloS one 4(12):e7992
Bao Z, Li X, Zan X, Shen L, Ma R, Liu W (2016) Signalling pathway impact analysis based on the strength of interaction between genes. IET Syst Biol 10(4):147–152
Bornholdt S (2005) Less is more in modeling large genetic networks. Science 310(5747):449–451
Buck V, Ng S, Ruiz-Garcia AB, Papadopoulou K, Bhatti S, Samuel JM, Anderson M, Millar JB, McInerny CJ (2004) Fkh2p and sep1p regulate mitotic gene transcription in fission yeast. J Cell Sci 117(23):5623–5632
Chai LE, Loh SK, Low ST, Mohamad MS, Deris S, Zakaria Z (2014) A review on the computational approaches for gene regulatory network construction. Comput Biol Med 48:55–65
Chaos A, Aldana M, Espinosa-Soto C, de León BGP, Arroyo AG, Alvarez-Buylla ER (2006) From genes to flower patterns and evolution: dynamic models of gene regulatory networks. J Plant Growth 25(4):278–289
Chaves M, Laurent T (2018) Analysis tools for interconnected Boolean networks with biological applications. Front Physiol 9:1–18
Chaves M, Figueiredo D, Martins MA (2018) Boolean dynamics revisited through feedback interconnections. Nat Comput. https://doi.org/10.1007/s11047-018-9716-8
Cheng D, Qi H (2010) A linear representation of dynamics of boolean networks. Trans Autom Control 55(10):2251–2258
Cheng D, Qi H, Li Z (2011) Model construction of boolean network via observed data. IEEE Trans Neural Netw 22(4):525–536
Ching WK, Chen X, Tsing NK (2009) Generating probabilistic boolean networks from a prescribed transition probability matrix. IET Syst Biol 3(6):453–464
Davidich MI, Bornholdt S (2008) Boolean network model predicts cell cycle sequence of fission yeast. PloS one 3(2):e1672
Devloo V, Hansen P, Labbé M (2003) Identification of all steady states in large networks by logical analysis. Bull Math Biol 65(6):1025–1051
Dougherty ER, Pal R, Qian X, Bittner ML, Datta A (2010) Stationary and structural control in gene regulatory networks: basic concepts. Int J Syst Sci 41(1):5–16
Espinosa-Soto C, Padilla-Longoria P, Alvarez-Buylla ER (2004) A gene regulatory network model for cell-fate determination during arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. Plant Cell 16(11):2923–2939
Feng W, Yang SX, Wu H (2011) On delayed uncertain genetic regulatory networks: robust stability analysis. Int J Comput Math 88(12):2448–2463
Gadouleau M, Richard A, Fanchon E (2018) Reduction and fixed points of boolean networks and linear network coding solvability. IEEE Trans Inf Theory 62(5):2504–2519
Glass L, Kauffman SA (1973) The logical analysis of continuous, non-linear biochemical control networks. J Theor Biol 39(1):103–129
Grinstead CM, Snell JL (2012) Introduction to probability. American Math. Soc, Providence
Heidel J, Maloney J, Farrow C, Rogers J (2003) Finding cycles in synchronous boolean networks with applications to biochemical systems. Int J Bifurc Chaos 13(03):535–552
Huang S (1999) Gene expression profiling, genetic networks, and cellular states: an integrating concept for tumorigenesis and drug discovery. Int J Mol Med 77(6):469–480
Huang S (2006) Cell state dynamics and tumorigenesis in Boolean regulatory networks. Springer, Berlin
Kauffman SA (1969) Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol 22(3):437–467
Kauffman SA (1993) The origins of order: self-organization and selection in evolution. Oxford University Press, Oxford
Lee S, Ko J, Tan X, Patel I, Balkrishnan R, Chang J (2014) Markov chain modelling analysis of HIV/AIDS progression: a race-based forecast in the united states. Indian J Pharm Sci 76(2):107
Li F, Long T, Lu Y, Ouyang Q, Tang C (2004) The yeast cell-cycle network is robustly designed. Proc Natl Acad Sci U S Am 101(14):4781–4786
Li H, Wang Y (2017) Robust stability and stabilisation of boolean networks with disturbance inputs. Int J Syst Sci 48(4):750–756
Li B (2017) Graphical reduction of probabilistic boolean networks. In: 36th Chinese control conference (CCC), 2017. IEEE, pp 1430–1434
Mendoza L, Xenarios I (2006) A method for the generation of standardized qualitative dynamical systems of regulatory networks. Theor Biol Med Model 3(1):13
Mochizuki A (2005) An analytical study of the number of steady states in gene regulatory networks. J Theor Biol 236(3):291–310
Needham CJ, Bradford JR, Bulpitt AJ, Westhead DR (2007) A primer on learning in bayesian networks for computational biology. PLOS Comput Biol 3(8):e129
Novak B, Pataki Z, Ciliberto A, Tyson JJ (2001) Mathematical model of the cell division cycle of fission yeast. Chaos interdiscip J Nonlinear Sci 11(1):277–286
Pal R, Ivanov I, Datta A, Bittner ML, Dougherty ER (2005) Generating boolean networks with a prescribed attractor structure. Bioinformatics 21(21):4021–4025
Paulevé L, Richard A (2012) Static analysis of boolean networks based on interaction graphs: a survey. Electron Notes Theor Comput Sci 284:93–104
Rejc Z, Magdevska L, Trselic T, Osolin T, Mraz J, Pavliha E, Zimic N, Cvitanovic T, Rozman D, Moskon M, Mraz M (2017) Computational modelling of genome-scale metabolic networks and its application to CHO cell cultures. Comput Biol Med 88:150–160
Remy É, Ruet P, Thieffry D (2008) Graphic requirements for multistability and attractive cycles in a boolean dynamical framework. Adv Appl Math 41(3):335–350
Rout RK, Choudhury PP, Sahoo S, Ray C (2015) Partitioning 1-variable boolean functions for various classification of n-variable boolean functions. Int J Comput Math 92(10):2066–2090
Rout RK, Pal Choudhury P, Sahoo S (2013) Classification of boolean functions where affine functions are uniformly distributed. J Discrete Math 2013:1–12
Rosenblueth DA, Muñoz S, Carrillo M, Azpeitia E (2014) Inference of Boolean networks from gene interaction graphs using a SAT solver. In: International conference on algorithms for computational biology. Springer, Cham, pp 235–246
Saadatpour A, Albert R, Reluga TC (2013) A reduction method for boolean network models proven to conserve attractors. SIAM J Appl Dyn Syst 12(4):1997–2011
Sahoo S, Choudhury PP, Chakraborty M (2008) Characterization of any non-linear boolean function using a set of linear operators. arXiv preprint arXiv:0808.1641
Seixas FL, Zadrozny B, Laks J, Conci A, Saade DCM (2014) A bayesian network decision model for supporting the diagnosis of dementia, Alzheimer's disease and mild cognitive impairment. Comput Biol Med 51:140–158
Shmulevich I, Dougherty ER (2010) Probabilistic Boolean networks: the modeling and control of gene regulatory networks. SIAM, Auckland
Sipari P (1991) Structured system models part 2. Directed graphs and boolean matrices. Int J Syst Sci 22(6):1071–1092
Thieffry D (2007) Dynamical roles of biological regulatory circuits. Brief Bioinform 8(4):220–225
Thomas R, d’Ari R (1990) Biological feedback. CRC Press, Boca Raton
Trepode NW, de Farias CR, Barrera J (2013) A pattern-oriented specification of gene network inference processes. Comput Biol Med 43(10):1415–1427
Veliz-Cuba A (2011) Reduction of boolean network models. J Theor Biol 289:167–172
Vichniac GY (1990) Boolean derivatives on cellular automata. Physica D 45(1–3):63–74
Wu CH, Sahoo D, Arvanitis C, Bradon N, Dill DL, Felsher DW (2013) Correction: combined analysis of murine and human microarrays and chip analysis reveals genes associated with the ability of myc to maintain tumorigenesis. PLoS Genet 9(10):1–16
Xiao M, Zheng WX, Cao J (2014) Stability and bifurcation of genetic regulatory networks with small rnas and multiple delays. Int J Comput Math 91(5):907–927
Zhang F, Hu Y, Jia Y, Xie M (2012) New constructions of balanced boolean functions with high nonlinearity and optimal algebraic degree. Int J Comput Math 89(10):1319–1331
Zhou Y, Xie M, Xiao G (2009) On cross-correlation properties of boolean functions. In: Fourth international conference on communications and networking in China, 2009. ChinaCOM 2009. IEEE, pp 1–5
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix: Boolean functions (BFs) are responsible to generate IGs having positive/negative edges only from 2 to 4-variable
Appendix: Boolean functions (BFs) are responsible to generate IGs having positive/negative edges only from 2 to 4-variable
Rights and permissions
About this article
Cite this article
Rout, R.K., Maity, S.P., Choudhury, P.P. et al. Analysis of Boolean functions based on interaction graphs and their influence in system biology. Neural Comput & Applic 32, 7803–7821 (2020). https://doi.org/10.1007/s00521-019-04102-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-019-04102-2