Abstract
An electrochemical method is described for ultrasensitive determination of protein tyrosine kinase-7 (PTK7). It is based on (a) the use of positively charged gold nanoparticles (AuNPs) and negatively charged graphene oxide (GO), and (b) of toehold-mediated strand displacement amplification. A hairpin probe 2 (HP2) containing the sgc8 aptamer was used to modify a glassy carbon electrode (GCE). Its hairpin structure is opened in the presence of PTK7 to form the PTK7-HP2 complex. The exposed part of HP2 partly hybridizes with hairpin probe 1 (HP1) that was immobilizing on the AuNPs and GO modified GCE. On addition of the hairpin probe 3 that was labeled with the redox probe Methylene Blue (MB-HP3), toehold-mediated strand displacement occurs due to complementary hybridization of HP1 with MB-HP3. This causes the release of PTK7-HP2 into the solution and makes it available for the next reaction. Under optimal conditions, PTK7 can be quantified by voltammetry (typically performed at −0.18 V) with a detection limit of 1.8 fM. The assay possesses high selectivity for PTK7 due to the employment of the aptamer. It was successfully applied to the determination of PTK7 in the debris of malignant melanoma A375 cells.

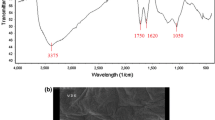
Schematic representation of the enzyme-free electrochemical sensor for ultrasensitive determination of protein tyrosine kinase-7 (PTK7) based on the toehold-mediated strand displacement reaction amplification on gold nanoparticles and graphene oxide.




Similar content being viewed by others
References
Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C, Croix BS (2001) A phosphatase associated with metastasis of colorectal cancer. Science 294:1343–1346
Kampen KR (2011) Membrane proteins: the key players of a cancer cell. J Membr Biol 242:69–74
Arinaminpathy Y, Khurana E, Engelman DM, Gerstein MB (2009) Computational analysis of membrane proteins: the largest class of drug targets. Drug Discov Today 14:1130–1135
Grimm D, Bauer J, Pietsch J, Infanger M, Eucker J, Eilles C, Schoenberger J (2011) Diagnostic and therapeutic use of membrane proteins in cancer cells. Curr Med Chem 18:176–190
Mothes W, Heinrich SU, Graf R, Nilsson I, Von HG, Brunner J, Rapoport TA (1997) Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell 89:523–533
Brown DM, Ruoslahti E (2004) A cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell 5:365–374
Hu MY, Li ZZ, Guo CP, Wang MH, He LH, Zhang ZH (2019) Hollow core-shell nanostructured MnO2/Fe2O3 embedded within amorphous carbon nanocomposite as sensitive bioplatform for detecting protein tyrosine kinase-7. Appl Surf Sci 489:13–24
Wagner G, Peradziryi H, Wehner P, Borchers A (2010) Plexin A1 interacts with PTK7 and is required for neural crest migration. Biochem Biophys Res Commun 402:402–407
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Shangguan DH, Li Y, Tang ZW, Cao ZHC, Chen HW, Mallikaratchy P, Sefah K, Yang CYJ, Tan WH (2006) Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A 103:11838–11843
Li L, Wang Q, Feng J, Tong LL, Tang B (2014) Highly sensitive and homogeneous detection of membrane protein on a single living cell by aptamer and nicking enzyme assisted signal amplification based on microfluidic droplets. Anal Chem 86:5101–5107
Lin S, Gao W, Tian Z, Yang C, Lu L, Mergny JL, Leung CH, Ma DL (2015) Luminescence switch-on detection of protein tyrosine kinase-7 using a G-quadruplex selective probe. Chem Sci 6:4284–4290
Yin JJ, He XX, Wang KM, Qing ZH, Wu X, Shi H, Yang XH (2011) One-step engineering of silver nanoclusters-aptamer assemblies as luminescent labels to target tumor cells. Nanoscale 4:110–112
Miao XM, Li ZB, Zhu AH, Feng ZZ, Tian J, Peng X (2016) Ultrasensitive electrochemical detection of protein tyrosine kinase-7 by gold nanoparticles and methylene blue assisted signal amplification. Biosens Bioelectron 83:39–44
Zheng J, Li NX, Li CR, Wang XX, Liu YC, Mao GB, Ji XH, He ZK (2018) A nonenzymatic DNA nanomachine for biomolecular detection by target recycling of hairpin DNA cascade amplification. Biosens Bioelectron 107:40–46
Li YR, Chang YY, Yuan R, Chai YQ (2018) Highly efficient target recycling-based netlike Y-DNA for regulation of electrocatalysis towards methylene blue for sensitive DNA detection. ACS Appl Mater Interfaces 10:25213–25218
Chen HG, Ren W, Jia J, Feng J, Gao ZF, Li NB, Luo HQ (2016) Fluorometric detection of mutant DNA oligonucleotide based on toehold strand displacement-driving target recycling strategy and exonuclease III-assisted suppression. Biosens Bioelectron 77:40–45
Ding W, Deng W, Zhu H, Liang HJ (2013) Metallo-toeholds: controlling DNA strand displacement driven by Hg(II) ions. Chem Commun 49:9953–9955
Miao XM, Cheng ZY, Ma HY, Li ZB, Xue N, Wang P (2018) Label-free platform for microRNA detection based on the fluorescence quenching of positively charged gold nanoparticles to silver nanoclusters. Anal Chem 90:1098–1103
Qu LL, Wang N, Xu H, Wang WP, Liu Y, Kuo LD, Yadav TP, Wu JJ, Joyner J, Song YH, Li HT, Lou J, Vajtai R, Ajayan PM (2017) Gold nanoparticles and g-C3N4-intercalated graphene oxide membrane for recyclable surface enhanced raman scattering. Adv Funct Mater 27:1701714
Easty DJ, Mitchell PJ, Patel K, Florenes VA, Spritz RA, Bennett DC (1997) Loss of expression of receptor tyrosine kinase family genes PTK7 and SEK in metastatic melanoma. Int J Cancer 71:1061–1065
Acknowledgements
This work was supported by the Natural Science Foundation of Xuzhou City (KC18140), and the project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest about this article.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 537 kb)
Rights and permissions
About this article
Cite this article
Li, Z., Zhou, Z., Xue, N. et al. Electrochemical aptamer-based determination of protein tyrosine kinase-7 using toehold-mediated strand displacement amplification on gold nanoparticles and graphene oxide. Microchim Acta 186, 720 (2019). https://doi.org/10.1007/s00604-019-3849-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3849-z