Abstract
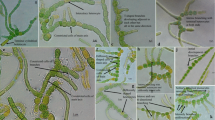
Two strains of true branched heterocytous cyanobacteria, SA33 and SA16, isolated from dried paddy fields of Mazandaran, Iran, were studied using morphological, ecological, and molecular tools. Morphological characterization of the strains indicated them to be commonly showing profuse T-type branching, large irregularly shaped cells of main filament and tapering branches. Strain SA33 showed development of interesting biseriate filaments with unilateral branching, while strain SA16 showed uniseriate filaments with bilateral branching. Ecological examination of the localities consisted of the assessment of the habitat and correlations with the overall environment of the habitats. 16S rRNA gene-based molecular and phylogenetic assessment gave unique positioning to both the strains separated with good bootstrap support from rest of the members of the true branched clade. Full length sequencing of the 16S–23S ITS region and folding of the secondary structures gave interesting secondary structures and comparison with the closely related sequences clearly indicated the secondary structures of both the strains to be unique. All the results indicated the strains to be members of a morphologically cryptic but phylogenetically distinct unknown genus of cyanobacteria. Comprehensive evaluation of all the findings and comparative assessment of previous studies indicate that SA33 and SA16 are indeed two new species of a new genus of true branched cyanobacteria. In accordance with the International Code of Nomenclature for algae, fungi and plants, we propose the name of the new genus as Neowestiellopsis with the names of the species being Neowestiellopsis persica and Neowestiellopsis bilateralis.




Similar content being viewed by others
References
Bagchi SN, Dubey N, Singh P (2017) Phylogenetically distant clade of Nostoc-like taxa with the description of Aliinostoc gen. nov. and Aliinostoc morphoplasticum sp. nov. Int J Syst Evol Microbiol 67:3329–3338. https://doi.org/10.1099/ijsem.0.002112
Bourrelly P (1970) Les algues d’eau douce. Initiation à la systématique, Tome III: Les Algues bleues et rouges, Les Eugléniens, Peridiniens et Cryptomonadines. Boubée & Cie, Paris
Dagan T, Roettger M, Stucken K, Landan G, Koch R, Major P, Gould SB, Goremykin VV, Rippka R, Marsac NTD, Gugger M, Lockhart PJ, Allen JF, Brune I, Maus I, Pühler A, Martin WF (2013) Genomes of stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol Evol 5:31–44. https://doi.org/10.1093/gbe/evs117
Desikachary TV (1959) Cyanophyta. ICAR monographs on algae, New Delhi
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucl Acids Res 17:7843–7853. https://doi.org/10.1093/nar/17.19.7843
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Molec Evol 17:368–376. https://doi.org/10.1007/BF01734359
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.2307/2408678
Fewer D (2001) Molecular evidence for the antiquity of group I introns interrupting transfer RNA genes in cyanobacteria. MSc Thesis, Universität zu Göttingen, Göttingen
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specified tree topology. Syst Zool 20:406–416. https://doi.org/10.2307/2412116
Genuário DB, Vaz GMV, Hentschke GS, Anna CLS, Fiore MF (2015) Halotia gen. nov., a phylogenetically and physiologically coherent cyanobacterial genus isolated from marine coastal environments. Int J Syst Evol Microbiol 65:663–675. https://doi.org/10.1099/ijs.0.070078-0
Gkelis S, Rajaniemi P, Vardaka E, Moustaka-Gouni M, Lanaras T, Sivonen K (2005) Limnothrix redekei (van goor) Meffert (cyanobacteria) strains from lake Kastoria, Greece form a separate phylogenetic group. Microbiol Ecol 49:176–182. https://doi.org/10.1007/s00248-0032030-7
Głowacka J, Szefel-Markowska M, Waleron M, Łojkowska E, Waleron K (2011) Detection and identification of potentially toxic cyanobacteria in Polish water bodies. Acta Biochim Pol 58:321–333
Gugger MF, Hoffmann L (2004) Polyphyly of true branching cyanobacteria (Stigonematales). Int J Syst Evol Microbiol 54:349–357. https://doi.org/10.1099/ijs.0.02744-0
Hoffmann L, Komárek J, Kaštovský J (2005) System of cyanoprokaryotes (cyanobacteria) state. Algol Stud 117:95–115. https://doi.org/10.1127/1864-1318/2005/0117-0095
Hrouzek P, Lukešová A, Mareš J, Ventura S (2013) Description of the cyanobacterial genus Desmonostoc gen. nov. including D. muscorum comb. nov. as a distinct, phylogenetically coherent taxon related to the genus Nostoc. Fottea 13:201–213
Janet M (1941) Westiellopsis prolifica, gen. et sp. nov., a new member of the Stigonemataceae. Ann Bot (Oxford) 5:167–170
Jeeji-Bai N (1972) The genus Westiellopsis Janet. In: Desikachary TV (ed) Taxonomy and biology of blue-green algae. University of Madras, Madras, pp 62–74
Kaštovský J, Johansen JR (2008) Mastigocladus laminosus (Stigonematales, Cyanobacteria): phylogenetic relationship of strains from thermal springs to soil-inhabiting genera of the order and taxonomic implications for the genus. Phycologia 47:307–320. https://doi.org/10.2216/PH07-69.1
Kaštovský J, Berrendero-Gomez E, Hladil J, Johansen JR (2014) Cyanocohniella calida gen. nov. et spec. nov. (Cyanobacteria: Aphanizomenonaceae) a new cyanobacterium from the thermal springs from Karlovy Vary, Czech Republic. Phytotaxa 181:279–292. https://doi.org/10.11646/phytotaxa.181.5.3
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. https://doi.org/10.1099/ijs.0.038075-0
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Molec Evol 16:111–120. https://doi.org/10.1007/BF01731581
Komárek J (2013) Cyanoprokaryota 3. Heterocytous genera. In: Büdel B, Gärtner G, Krienitz L, Schagerl M (eds) Süβwasserflora von mitteleuropa/freshwater flora of Central Europe. Springer, Heidelberg, p 1130
Komárek J, Mareš J (2012) An update to modern taxonomy (2011) of freshwater planktic heterocytous cyanobacteria. Hydrobiologia 698:327–351. https://doi.org/10.1007/s10750-012-1027y
Komárek J, Kaštovský J, Mareš J, Johansen JR (2014) Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) using a polyphasic approach. Preslia 86:295–335
Komárek J, Komárková J, Ventura S, Kozlíková-Zapomělová E, Rejmánková E (2017) Taxonomic evaluation of cyanobacterial microflora from alkaline marshes of northern Belize. 3. Diversity of heterocytous genera. Nova Hedwigia 105:445–486. https://doi.org/10.1127/nova_hedwigia/2017/0425
Řeháková K, Johansen JR, Casamatta DA, Xuesong L, Vincent J (2007) Morphological and molecular characterization of selected desert soil cyanobacteria: three species new to science including Mojavia pulchra gen. et sp. nov. Phycologia 46:481–502. https://doi.org/10.2216/06-92.1
Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure culture of cyanobacteria. J Gen Microbiol 111:1–61. https://doi.org/10.1099/00221287-111-1-1
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molec Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Singh P, Singh SS, Mishra AK, Elster J (2013) Molecular phylogeny, population genetics and evolution of heterocystous cyanobacteria using nifH gene sequences. Protoplasma 250:751–764. https://doi.org/10.1007/s00709-012-0460-0
Singh P, Kaushik MK, Srivastava M, Mishra AK (2014) Phylogenetic analysis of heterocystous cyanobacteria (subsections IV and V) using highly iterated palindromes as molecular markers. Physiol Molec Biol Pl 20:331–342. https://doi.org/10.1007/s12298-014-0244-4
Singh P, Fatma A, Mishra AK (2015a) Molecular phylogeny and evogenomics of heterocystous cyanobacteria using rbcl gene sequence data. Ann Microbiol 65:799–807. https://doi.org/10.1007/s13213-014-0920-1
Singh P, Singh SS, Aboal M, Mishra AK (2015b) Decoding cyanobacterial phylogeny and molecular evolution using an evonumeric approach. Protoplasma 252:519–535. https://doi.org/10.1007/s00709-014-0699-8
Singh P, Dubey N, Bagchi SN (2017) Westiellopsis ramosa sp. nov., intensely branched species of Westiellopsis (Cyanobacteria) from freshwater habitat in Jabalpur, Madhya Pradesh, India. Pl Syst Evol 303:1239–1249. https://doi.org/10.1007/s00606-017-1434-7
Suradkar A, Villanueva C, Gaysina LA, Casamatta DA, Saraf A, Dighe G, Mergu R, Singh P (2017) Nostoc thermotolerans sp. nov., a soil-dwelling species of Nostoc (Cyanobacteria). Int J Syst Evol Microbiol 67:1296–1305. https://doi.org/10.1099/ijsem.0.001800
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molec Biol Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Zucker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucl Acids Res 31:3406–3415. https://doi.org/10.1093/nar/gkg595
Acknowledgements
We thank the Director NCCS for providing the necessary facilities and encouragement. PS is thankful to the Department of Science and Technology (Department of Science and Technology), India for the project YSS/2014/000879. A part of the work was supported by the Department of Biotechnology (Department of Biotechnology; Grant No. BT/Coord.II/01/032016), Government of India, under the project ‘Establishment of Centre of Excellence for National Centre for Microbial Resource (NCMR)’. SK was the recipient of the NAM S&T Centre Research Training Fellowship for Developing Country Scientists (RTF-DCS) 2016–17 and thanks the NAM Centre for providing research fellowship. The authors thank Prof. Aharon Oren for all the help in the Latin nomenclature and etymology. Thanks to Archana Suradkar and Aniket Saraf for help throughout the morphological analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling editor: Karol Marhold.
Accession Numbers generated through this study: MF066911-12, MF115981-86.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
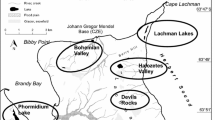
Online Resource 1. Physicochemical parameters and climate data of the habitat from which Westiellopsis persica SA33 and W. bilateralis SA16 originated.
Online Resource 2. 16S rRNA gene alignment files of the strains SA16 and SA33 along with the rest of the strains in the phylogenetic analysis.
Online Resource 3. Phylogenetic tree based on the rbcL gene of the strains Neowestiellopsis persica SA33 and N. bilateralis SA16 with the bootstrap values representing NJ, ML and MP, respectively.
Online Resource 4. rbcL gene alignment files of the strains SA16 and SA33 along with the rest of the strains in the phylogenetic analysis.
Online Resource 5. Phylogenetic tree based on the rpoC1 gene of the strains Neowestiellopsis persica SA33 and N. bilateralis SA16 with the bootstrap values representing NJ, ML, and MP, respectively.
Online Resource 6. rpoC1 gene alignment files of the strains SA16 and SA33 along with the rest of the strains in the phylogenetic analysis.
Online Resource 7. Phylogenetic tree based on the tufA gene of the strains Neowestiellopsis persica SA33 and N. bilateralis SA16 with the bootstrap values representing NJ, ML, and MP, respectively.
Online Resource 8. tufA gene alignment files of the strains SA16 and SA33 along with the rest of the strains in the phylogenetic analysis.
Online Resource 9. p-distance values of all the strains included in the ITS analysis.
Rights and permissions
About this article
Cite this article
Kabirnataj, S., Nematzadeh, G.A., Talebi, A.F. et al. Neowestiellopsis gen. nov, a new genus of true branched cyanobacteria with the description of Neowestiellopsis persica sp. nov. and Neowestiellopsis bilateralis sp. nov., isolated from Iran. Plant Syst Evol 304, 501–510 (2018). https://doi.org/10.1007/s00606-017-1488-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-017-1488-6