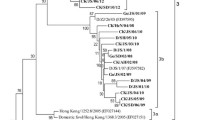
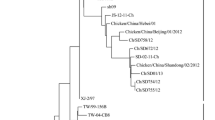
Abstract
Given the global evolutionary dynamics of Newcastle disease viruses (NDVs), it is imperative to continue extensive surveillance, routine monitoring and characterization of isolates originating from natural reservoirs (waterfowls). In this report, we isolated and characterized two virulent NDV strains from clinically healthy mallard (Anas platyrhynchos). Both isolates had a genome of 15,192 nucleotides encoding six genes in an order of 3´-NP-P-M-F-HN-L-5´. The biological characteristics (mean death time: 49.5-50 hr, EID50108.5 ml−1) and presence of a typical cleavage site in the fusion (F) protein (112R-R-Q-K-R↓F117) confirmed the velogenic nature of these isolates. Phylogenetic analysis classified both isolates as members of genotype VII within class-II. Furthermore, based upon the hypervariable region of the F gene (375 nt), isolates showed clustering within sub-genotype VIIi. Similarity index and parallel comparison revealed a higher nucleotide divergence from commonly used vaccine strains; LaSota (21%) and Mukteswar (17%). A comparative residues analysis with representative strains of different genotypes, including vaccine strains, revealed a number of substitutions at important structural and functional domains within the F and hemagglutinin-neuraminidase (HN) proteins. Together, the results highlight consistent evolution among circulating NDVs supporting extensive surveillance of the virus in waterfowl to better elucidate epidemiology, evolutionary relationships and their impacts on commercial and backyard poultry.




Similar content being viewed by others
References
Akbar M, Mushtaq-Ul-Hassan M, Mahmood-Ul-Hassan M, Hassan M (2009) Waterfowl diversity at Chashma barrage (Wildlife Sanctuary Mianwali) and Marala headworks (Game Reserve Sialkot), Pakistan during 1996–2005. Int J Agric Biol 11:188–192
Akhtar S, Muneer MA, Muhammad K, Tipu MY, Rabbani M, Shabbir MZ (2016) Genetic characterization and phylogeny of pigeon paramyxovirus isolate (PPMV-1) from Pakistan. SpringerPlus 5:1295
Aldous E, Alexander D (2001) Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathol 30:117–128
Alexander D (1995) The epidemiology and control of avian influenza and Newcastle disease. J Comp Pathol 112:105–126
Alexander D, Senne D (2008) Newcastle disease, other avian paramyxoviruses, and pneumovirus infections. Diseases of poultry, 12th edn. Blackwell, Ames, pp 75–98
Bahl A, Pomeroy B, Mangundimedjo S, Easterday B (1977) Isolation of type A influenza and Newcastle disease viruses from migratory waterfowl in the Mississippi flyway. J Am Vet Med Assoc 171:949–951
Ballagi-Pordany A, Wehmann E, Herczeg J, Belak S, Lomniczi B (1996) Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Adv Virol 141:243–261
Bilal ESA, Elnasri IM, Alhassan AM, Khalifa KA, Elhag JI, Ahmed SO (2014) Biological pathotyping of newcastle disease viruses in Sudan 2008–2013. J Vet Med 2014:1–4
Calain P, Roux L (1993) The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol 67:4822–4830
Collins M, Bashiruddin J, Alexander D (1993) Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity. Adv Virol 128:363–370
OIE (2012) Manual of diagnostic tests and vaccines for terrestrial animals: mammals, birds and bees, Biological Standards Commission. World Organization for Animal Health, Paris, pp 1–19
Connaris H, Takimoto T, Russell R, Crennell S, Moustafa I, Portner A, Taylor G (2002) Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J Virol 76:1816–1824
Crennell S, Takimoto T, Portner A, Taylor G (2000) Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Mol Biol 7:1068
Czeglédi A, Ujvári D, Somogyi E, Wehmann E, Werner O, Lomniczi B (2006) Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res 120:36–48
Dimitrov KM, Bolotin V, Muzyka D, Goraichuk IV, Solodiankin O, Gerilovych A, Stegniy B, Goujgoulova GV, Silko NY, Pantin-Jackwood MJ (2016) Repeated isolation of virulent Newcastle disease viruses of sub-genotype VIId from backyard chickens in Bulgaria and Ukraine between 2002 and 2013. Adv Virol 161:3345–3353
Dimitrov KM, Ramey AM, Qiu X, Bahl J, Afonso CL (2016) Temporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus). Infection, genetics and evolution 39:22–34
Fiebig L, Soyka J, Buda S, Buchholz U, Dehnert M, Haas W (2011) Avian influenza A (H5N1) in humans: new insights from a line list of World Health Organization confirmed cases, September 2006 to August 2010
Friend M, Trainer D (1970) Serologic evidence of Newcastle disease in captive mallards and swans. J Wildl Dis 6:130–135
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic acids symposium series. [London]: Information Retrieval Ltd., c1979–c2000., pp 95–98
Health WOfA (2012) Manual of diagnostic tests and vaccines for terrestrial animals: mammals, birds and bees. Biological Standards Commission, World Organization for Animal Health Paris, France
Iorio RM, Syddall RJ, Glickman RL, Kiel AM, Sheehan JP, Bratt MA (1989) Identification of amino acid residues important to the neuraminidase activity of the HN glycoprotein of Newcastle disease virus. Virology 173:196–204
Iorio RM, Syddall RJ, Sheehan JP, Bratt MA, Glickman RL, Riel AM (1991) Neutralization map of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus: domains recognized by monoclonal antibodies that prevent receptor recognition. J Virol 65:4999–5006
Iorio RM, Field GM, Sauvron JM, Mirza AM, Deng R, Mahon PJ, Langedijk JP (2001) Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J Virol 75:1918–1927
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kim LM, King DJ, Curry PE, Suarez DL, Swayne DE, Stallknecht DE, Slemons RD, Pedersen JC, Senne DA, Winker K (2007) Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J Virol 81:12641–12653
Kolakofsky D, Roux L, Garcin D, Ruigrok RW (2005) Paramyxovirus mRNA editing, the ‘rule of six’and error catastrophe: a hypothesis. J Gen Virol 86:1869–1877
Lamb RA, Paterson RG, Jardetzky TS (2006) Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 344:30–37
Lee E-K, Jeon W-J, Kwon J-H, Yang C-B, Choi K-S (2009) Molecular epidemiological investigation of Newcastle disease virus from domestic ducks in Korea. Vet Microbiol 134:241–248
Lindh E, Ek-Kommonen C, Väänänen V-M, Alasaari J, Vaheri A, Vapalahti O, Huovilainen A (2012) Molecular epidemiology of outbreak-associated and wild waterfowl-derived Newcastle disease virus strains in Finland, including a novel class I genotype. J Clin Microbiol 50:366423673
Liu X, Wan H, Ni X, Wu Y, Liu W (2003) Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose flocks in some regions of China during 1985–2001. Adv Virol 148:1387–1403
Maminiaina OF, Gil P, Briand F-X, Albina E, Keita D, Andriamanivo HR, Chevalier V, Lancelot R, Martinez D, Rakotondravao R (2010) Newcastle disease virus in Madagascar: identification of an original genotype possibly deriving from a died out ancestor of genotype IV. PLoS One 5:e13987
Mayo M (2002) A summary of taxonomic changes recently approved by ICTV. Adv Virol 147:1655–1656
Miller PJ, Dimitrov KM, Williams-Coplin D, Peterson MP, Pantin-Jackwood MJ, Swayne DE, Suarez DL, Afonso CL (2015) International biological engagement programs facilitate Newcastle disease epidemiological studies. Front Public Health 3:235
Miller PJ, Haddas R, Simanov L, Lublin A, Rehmani SF, Wajid A, Bibi T, Khan TA, Yaqub T, Setiyaningsih S (2015) Identification of new sub-genotypes of virulent Newcastle disease virus with potential panzootic features. Infect Genet Evol 29:216–229
Munir M, Linde A-M, Zohari S, Ståhl K, Baule C, Holm K, Engström B, Berg M (2010) Complete genome analysis of an avian paramyxovirus type 1 strain isolated in 1994 from an asymptomatic black-headed gull (Larus ridibundus) in southern Sweden. Avian Dis 54:923–930
Munir M, Abbas M, Khan MT, Zohari S, Berg M (2012) Genomic and biological characterization of a velogenic Newcastle disease virus isolated from a healthy backyard poultry flock in 2010. Virol J 9:46
Pantin-Jackwood MJ, Miller PJ, Spackman E, Swayne DE, Susta L, Costa-Hurtado M, Suarez DL (2014) Role of poultry in the spread of novel H7N9 influenza virus in China. J Virol 88:5381–5390
Perozo F, Marcano R, Afonso CL (2012) Biological and phylogenetic characterization of a genotype VII Newcastle disease virus from Venezuela: efficacy of field vaccination. J Clin Microbiol 50:1204–1208
Radwan M, Darwish S, El-Sabagh I, El-Sanousi A, Shalaby M (2013) Isolation and molecular characterization of Newcastle disease virus genotypes II and VIId in Egypt between 2011 and 2012. Virus Genes 47:311–316
Reed LJ, Muench H (1938) A simple method of estimating fifty per cent endpoints 12. Am J Epidemiol 27:493–497
Römer-Oberdörfer A, Werner O, Veits J, Mebatsion T, Mettenleiter TC (2003) Contribution of the length of the HN protein and the sequence of the F protein cleavage site to Newcastle disease virus pathogenicity. J Gen Virol 84:3121–3129
Sakai K, Sakabe G, Tani O, Watanabe Y, Jahangir A, Nakamura M, Takehara K (2007) Characterization of Newcastle disease virus isolated from northern pintail (Anas acuta) in Japan. J Vet Med Sci 69:1307–1311
Samal S (2012) Role of fusion protein in Newcastle disease virus pathogenesis. University of Maryland, College Park
Shabbir MZ, Malik AN, Wajid A, Rehmani SF, Munir M (2012) Newcastle disease virus: disease appraisal with global and Pakistan perspectives. Virus Genes 46:309–315
Shabbir MZ, Malik AN, Wajid A, Rehmani SF, Munir M (2013) Newcastle disease virus: disease appraisal with global and Pakistan perspectives. J Infect Molecul Biol 1:52–57
Shengqing Y, Kishida N, Ito H, Kida H, Otsuki K, Kawaoka Y, Ito T (2002) Generation of velogenic Newcastle disease viruses from a nonpathogenic waterfowl isolate by passaging in chickens. Virology 301:206–211
Stanislawek W, Wilks C, Meers J, Horner G, Alexander D, Manvell R, Kattenbelt J, Gould A (2002) Avian paramyxoviruses and influenza viruses isolated from mallard ducks (Anas platyrhynchos) in New Zealand. Adv Virol 147:1287–1302
Stone-Hulslander J, Morrison TG (1999) Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J Virol 73:3630–3637
Suarez D, Miller P, Koch G, Mundt E, Rautenschlein S (2012) Newcastle disease, other Avian paramyxoviruses, and avian metaneumovirus infections. In: Swayne DE, Glisson JR, McDougald LR, Nolan LK, Suarez D, Nair V (eds) Diseases of Poultry, 13th edn. Wiley-Blackwell, Iowa
Takakuwa H, Ito T, Takada A, Okazaki K, Kida H (1998) Potentially virulent Newcastle disease viruses are maintained in migratory waterfowl populations. Jpn J Vet Res 45:207–215
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tsai H-J, Chang K-H, Tseng C-H, Frost KM, Manvell RJ, Alexanderc DJ (2004) Antigenic and genotypical characterization of Newcastle disease viruses isolated in Taiwan between 1969 and 1996. Vet Microbiol 104:19–30
Umali DV, Ito H, Suzuki T, Shirota K, Katoh H, Ito T (2013) Molecular epidemiology of Newcastle disease virus isolates from vaccinated commercial poultry farms in non-epidemic areas of Japan. Virol J 10:330
Uyeki TM (2009) Human infection with highly pathogenic avian influenza A (H5N1) virus: review of clinical issues. Clin Infect Dis 49:279–290
Wajid A, Dimitrov KM, Wasim M, Rehmani SF, Basharat A, Bibi T, Arif S, Yaqub T, Tayyab M, Ababneh M (2017) Repeated isolation of virulent Newcastle disease viruses in poultry and captive non-poultry avian species in Pakistan from 2011 to 2016. Prev Vet Med 142:1–6
Xiao S, Paldurai A, Nayak B, Samuel A, Bharoto EE, Prajitno TY, Collins PL, Samal SK (2012) Complete genome sequences of Newcastle disease virus strains circulating in chicken populations of Indonesia. J Virol 86:5969–5970
Yang C-Y, Shieh HK, Lin Y-L, Chang P-C (1999) Newcastle disease virus isolated from recent outbreaks in Taiwan phylogenetically related to viruses (genotype VII) from recent outbreaks in western Europe. Avian Dis 43:125–130
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8
Yusoff K, Nesbit M, McCartney H, Meulemans G, Alexander D, Collins M, Emmerson P, Samson A (1989) Location of neutralizing epitopes on the fusion protein of Newcastle disease virus strain Beaudette C. J Gen Virol 70:3105–3109
Acknowledgements
We acknowledge team members of Pak-UK collaborative research project entitled “Combating avian influenza through systemic analysis of antigenic drift, genetic variation and development of diagnostic tools and vaccines” (Grant reference: BB/L018853 funded by BBSRC/UK) for providing study isolates and necessary data.
Author information
Authors and Affiliations
Contributions
Conceived and designed the work: MH, TY, JN, MZS. Performed the laboratory procedures and relevant methods: MH, AR, MM, JN, TY, TS, MZS. Data analysis: MH, AR, MM, MZS. Necessary laboratory resources and consumables: TY, WS, JN, MZS. Draft writing and editing: MH, AR, MM, MZS.
Corresponding author
Ethics declarations
Conflict of interest
All authors declared no conflict of interest with data presented in this manuscript.
Ethics statement
All animal handling and sample processing procedures were carried out in strict accordance of institutional guidelines and regulations related to Animal Welfare and Health. The used procedures were approved by the Ethical Review Committee for use of Laboratory Animals (ERCULA) of the University of Veterinary and Animal Sciences, Lahore, Pakistan (Permit Number: ORIC/DR-70).
Additional information
Handling Editor: Zhenhai Chen.
Rights and permissions
About this article
Cite this article
Habib, M., Yaqub, T., Nazir, J. et al. Genomic and biological characterization of Newcastle disease viruses isolated from migratory mallards (Anas platyrhynchos). Arch Virol 163, 2179–2188 (2018). https://doi.org/10.1007/s00705-018-3840-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3840-8