Abstract
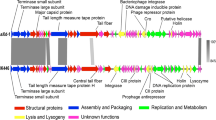
We present here the results of the analysis of the complete genome sequence of a lytic bacteriophage, vB_ButM_GuL6, which is the first virus isolated from Buttiauxella. Electron microscopy revealed that vB_ButM_GuL6 belongs to the family Myoviridae, order Caudovirales. The genome of vB_ButM_GuL6 is a linear, circularly permuted 178,039-bp dsDNA molecule with a GC content of 43.4%. It has been predicted to contain 282 protein-coding genes and two tRNA genes, tRNA-Met and tRNA-Gly. Using bioinformatics approaches, 99 (36%) of the vB_ButM_GuL6 genes were assigned a putative function. Genome-wide comparisons and phylogenetic analysis indicated that vB_ButM_GuL6 represents a new species of the subfamily Tevenvirinae and is most closely related to Escherichia virus RB43. These phages, together with Cronobacter phages Miller, CfP1, and IME-CF2, likely form a new genus within the subfamily Tevenvirinae.


Similar content being viewed by others
Data availability
The complete genome sequence of Buttiauxella phage vB_ButM_GuL6 was deposited in the GenBank database under the accession number MT334653.
References
Ferragut C, Izard D, Gavini F, Lefebvre B, Leclerc H (1981) Buttiauxella, a new genus of the family Enterobacteraceae. Zentralbl Bakteriol Mikrobiol Hyg A 2:33–44. https://doi.org/10.1016/S0721-9571(81)80016-6
Kämpfer P (2015) Buttiauxella. In: Bergey’s manual of systematics of Archaea and Bacteria, pp 1–16. https://doi.org/10.1002/9781118960608.gbm01139
Batinovic S, Wassef F, Knowler SA, Rice DTF, Stanton CR, Rose J, Tucci J, Nittami T, Vinh A, Drummond GR, Sobey CG, Chan HT, Seviour RJ, Petrovski S, Franks AE (2019) Bacteriophages in natural and artificial environments. Pathogens 8:100. https://doi.org/10.3390/pathogens8030100
Carlson K (2005) Appendix: working with bacteriophages: common techniques and methodological approaches. In: Kutter E, Sulakvelidze A (eds) Bacteriophages: biology and applications. CRC Press, London
Kaliniene L, Truncaitė L, Šimoliūnas E et al (2018) Molecular analysis of the low-temperature Escherichia coli phage vB_EcoS_NBD2. Arch Virol 163:105–114. https://doi.org/10.1007/s00705-017-3589-5
Carlson K, Miller E (1994) Experiments in T4 genetics. In: Karam JD (ed) Bacteriophage 797 T4. ASM Press, Washington DC, pp 419–483
Lowe TM, Eddy SR (1997) TRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. https://doi.org/10.1093/nar/25.5.0955
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. https://doi.org/10.1093/nar/gki408
McNair K, Bailey BA, Edwards RA (2012) PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 28:614–618. https://doi.org/10.1093/bioinformatics/bts014
Meier-Kolthoff JP, Goker M (2017) VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics (Oxford, England) 33:3396–3404. https://doi.org/10.1093/bioinformatics/btx440
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Funding
This study was supported by the Research Council of Lithuania (project no. SIT-7/2015).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Noreika, A., Meškys, R., Lazutka, J. et al. Complete genome sequence of Buttiauxella phage vB_ButM_GuL6. Arch Virol 165, 2685–2687 (2020). https://doi.org/10.1007/s00705-020-04780-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04780-7