Abstract
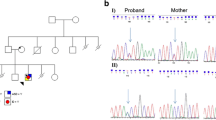
Pathogenic sequence variants in the IQ motif– and Sec7 domain–containing protein 2 (IQSEC2) gene have been confirmed as causative in the aetiopathogenesis of neurodevelopmental disorders (intellectual disability, autism) and epilepsy. We report on a case of a family with three sons; two of them manifest delayed psychomotor development and epilepsy. Initially proband A was examined using a multistep molecular diagnostics algorithm, including karyotype and array-comparative genomic hybridization analysis, both with negative results. Therefore, probands A and B and their unaffected parents were enrolled for an analysis using targeted “next-generation” sequencing (NGS) with a gene panel ClearSeq Inherited DiseaseXT (Agilent Technologies) and verification analysis by Sanger sequencing. A novel frameshift variant in the X-linked IQSEC2 gene NM_001111125.2:c.1813_1814del, p.(Asp605Profs*3) on protein level, was identified in both affected probands and their asymptomatic mother, having skewed X chromosome inactivation (XCI) (100:0). As the IQSEC2 gene is a known gene escaping from XCI in humans, we expect the existence of mechanisms maintaining the normal or enough level of the IQSEC2 protein in the asymptomatic mother. Further analyses may help to the characterization of the presented novel frameshift variant in the IQSEC2 gene as well as to elucidate the mechanisms leading to the rare asymptomatic phenotypes in females.


Similar content being viewed by others
Data availability
The microarray data obtained in the present study are available in the Array Express database (https://www.ebi.ac.uk/arrayexpress/) in the txt format under the accession number E-MTAB-8147. The NGS data analyzed in this study are available in the Array Express database (https://www.ebi.ac.uk/arrayexpress/) in fastq and bam formats under the accession number E-MTAB-8131. Sanger sequencing data, including the chromatograms of the probands and the parents, are stored in the Figshare online digital repository (https://doi.org/10.6084/m9.figshare.8667800.v1). The novel IQSEC2 gene variant NM_001111125.2:c.1813_1814del was submitted to Leiden Open Variation Database v.3.0 (Global Variome shared LOVD) under the accession number #0000629453 (https://databases.lovd.nl/shared/variants/0000629453). The variants and phenotypes were submitted to Leiden Open Variation Database v.3.0 (Global Variome shared LOVD). The novel IQSEC2 gene variant NM_001111125.2:c.1813_1814del is available under the accession number #0000629453 (https://databases.lovd.nl/shared/variants/0000629453). The BTD gene variant NM_000060.2:c.1330G>C is available under the accession number #0000629454 (https://databases.lovd.nl/shared/variants/0000629454).
References
Kochinke K, Zweier C, Nijhof B, Fenckova M, Cizek P, Honti F, Keerthikumar S, Oortveld MA, Kleefstra T, Kramer JM et al (2016) Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am J Hum Genet 98(1):149–164
Neri G, Schwartz CE, Lubs HA, Stevenson RE (2018) X-linked intellectual disability update 2017. Am J Med Genet A 176(6):1375–1388
Shoubridge C, Harvey RJ, Dudding-Byth T (2019) IQSEC2 mutation update and review of the female-specific phenotype spectrum including intellectual disability and epilepsy. Hum Mutat 40(1):5–24
Rogers EJ, Jada R, Schragenheim-Rozales K, Sah M, Cortes M, Florence M, Levy NS, Moss R, Walikonis RS, Palty R, Shalgi R, Lichtman D, Kavushansky A, Gerges NZ, Kahn I, Umanah GKE, Levy AP (2019) An IQSEC2 mutation associated with intellectual disability and autism results in decreased surface AMPA receptors. Front Mol Neurosci 12:43. https://doi.org/10.3389/fnmol.2019.00043
Zerem A, Haginoya K, Lev D, Blumkin L, Kivity S, Linder I, Shoubridge C, Palmer EE, Field M, Boyle J, Chitayat D, Gaillard WD, Kossoff EH, Willems M, Geneviève D, Tran-Mau-Them F, Epstein O, Heyman E, Dugan S, Masurel-Paulet A, Piton A', Kleefstra T, Pfundt R, Sato R, Tzschach A, Matsumoto N, Saitsu H, Leshinsky-Silver E, Lerman-Sagie T (2016) The molecular and phenotypic spectrum of IQSEC2-related epilepsy. Epilepsia 57(11):1858–1869
Wayhelova M, Oppelt J, Smetana J, Hladilkova E, Filkova H, Makaturova E, Nikolova P, Beharka R, Gaillyova R, Kuglik P (2019) Novel de novo frameshift variant in the ASXL3 gene in a child with microcephaly and global developmental delay. Mol Med Rep 20(1):505–512
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, Massouras A (2019) VarSome: the human genomic variant search engine. Bioinformatics 35(11):1978–1980
Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) Mutation Taster2: mutation prediction for deep-sequencing age. Nat Methods 11(4):361–362
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A et al (2016) The Ensembl Variant Effect Predictor. Genome Biol 17(1):122. https://doi.org/10.1186/s13059-016-0974-4
Coban-Akdemir Z, White JJ, Song X, Jhangiani SN, Faith JM, Gambin T, Bayram Y, Chinn IK, Karaca E, Punetha J et al (2018) Identifying genes whose mutant transcripts cause dominant disease traits by potential gain-of-function alleles. Am J Hum Genet 103(2):171–187
Lindeboom RGH, Vermeulen M, Lehner B, Supek F (2019) The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat Genet 51(11):1645–1651
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW (1992) Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen receptor gene correlates with X chromosome inactivation. Am J Hum Genet 51(6):1229–1239
Desai V, Donsante A, Swoboda KJ, Martensen M, Thompson J, Kaler SG (2011) Favorably skewed X-inactivation accounts for neurological sparing in female carriers of Menkes disease. Clin Genet 79(2):176–182
Kanaan SB, Onat OE, Balandraud N, Martin GV, Nelson JL, Azzouz DF, Auger I, Arnoux F, Martin M, Roudier J, Ozcelik T, Lambert NC (2016) Evaluation of X chromosome inactivation with respect to HLA genetic susceptibility in rheumatoid arthritis and systemic sclerosis. PLos One 11(6):e0158550. https://doi.org/10.1371/journal.pone.0158550
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424
Shoubridge C, Tarpey PS, Abidi F, Ramsden SL, Rujirabanjerd S, Murphy JA, Boyle J, Shaw M, Gardner A, Proos A, Puusepp H, Raymond FL, Schwartz CE, Stevenson RE, Turner G, Field M, Walikonis RS, Harvey RJ, Hackett A, Futreal PA, Stratton MR, Gécz J (2010) Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nat Genet 42(6):486–468
Mignot C, McMahon AC, Bar C, Campeau PM, Davidson C, Buratti J, Nava C, Jacquemont ML, Tallot M, Milh M et al (2019) IQSEC2-related encephalopathy in males and females: a comparative study including 37 novel patients. Genet Med 21(4):837–849
Kalscheuer VM, James VM, Himelright ML, Long P, Oegema R, Jensen C, Bienek M, Hu H, Haas SA, Topf M, Hoogeboom AJM, Harvey K, Walikonis R, Harvey RJ (2016) Novel missense mutation A789V in IQSEC2 underlies X-linked intellectual disability in the MRX78 family. Front Mol Neurosci 8:85. https://doi.org/10.3389/fnmol.2015.00085
Hinze SJ, Jackson MR, Lie S, Jolly L, Field M, Barry SC, Harvey RJ, Shoubridge C (2017) Incorrect dosage of IQSEC2, a known intellectual disability and epilepsy gene, disrupts dendritic spine morphology. Transl Psychiatry 7(5):e1110. https://doi.org/10.1038/tp.2017.81
Levy NS, Umanah GKE, Rogers EJ, Jada R, Lache O, Levy AP (2019) IQSEC2-associated intellectual disability and autism. Int J Mol Sci 20(12):E3038. https://doi.org/10.3390/ijms20123038
Ewans LJ, Field M, Zhu Y, Turner G, Leffler M, Dinger ME, Cowley MJ, Buckley MF, Scheffer IE, Jackson MR, Roscioli T, Shoubridge C (2017) Gonadal mosaicism of a novel IQSEC2 variant causing female limited intellectual disability and epilepsy. Eur J Hum Genet 25(6):763–767
Lindeboom RG, Supek F, Lehner B (2016) The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet 48(10):1112–1118
Kurosaki T, Maquat LE (2016) Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 129(3):461–467
Radley JA, O’Sullivan RBG, Turton SE, Cox H, Vogt J, Morton J, Jones E, Smithson S, Lachlan K, Rankin J et al (2019) Deep phenotyping of 14 new patients with IQSEC2 variants, including monozygotic twins of discordant phenotype. Clin Genet 95(4):496–506
Bittel DC, Theodoro MF, Kibiryeva N, Fischer W, Talebizadeh Z, Butler MG (2008) Comparison of X-chromosome inactivation patterns in multiple tissues from human females. J Med Genet 45(5):309–313
Li G, Zhang Z, Jin T, Liang H, Tu Y, Gong L, Chen Z, Gao G (2013) High frequency of the X-chromosome inactivation in young female patients with high-grade glioma. Diagn Pathol 8:101. https://doi.org/10.1186/1746-1596-8-101
Szelinger S, Malenica I, Corneveaux JJ, Siniard AL, Kurdoglu AA, Ramsey KM, Schrauwen I, Trent JM, Narayanan V, Huentelman MJ, Craig DW (2014) Characterization of X chromosome inactivation using integrated analysis of whole-exome and mRNA sequencing. PLoS One 9(12):e113036. https://doi.org/10.1371/journal.pone.0113036
Wong CC, Caspi A, Williams B, Houts R, Craig IW, Mill J (2011) A longitudinal twin study of skewed X-chromosome inactivation. PLoS One 6(3):e17873. https://doi.org/10.1371/journal.pone.0017873
Hatakeyama C, Anderson CL, Beever CL, Penaherrera MS, Brown CJ, Robinson WP (2004) The dynamics of X-inactivation skewing as women age. Clin Genet 66(4):327–332
Van den Veyver IB (2001) Skewed X inactivation in X-linked disorders. Semin Reprod Med 19(2):183–191
Shvetsova E, Sofronova A, Monajemi R, Gagalova K, Draisma HHM, White SJ, Santen GWE, Chuva de Sousa Lopes SM, Heijmans BT, van Meurs J et al (2019) Skewed X-inactivation is common in the general female population. Eur J Hum Genet 27(3):455–465
Barrie ES, Cottrell CE, Gastier-Foster J, Hickey SE, Patel AD, Santoro SL, Alfaro MP (2020) Genotype-phenotype correlation: inheritance and variant-type infer pathogenicity in IQSEC2 gene. Eur J Med Genet 63(3):103735. https://doi.org/10.1016/j.ejmg.2019.103735
Anderson CL, Brown CJ (1999) Polymorphic X-chromosome inactivation of the human TIMP1 gene. Am J Hum Genet 65(3):699–708
Happ HC, Carvill GL (2020) A 2020 view on the genetics of developmental and epileptic encephalopathies. Epilepsy Curr 20:90–96. https://doi.org/10.1177/1535759720906118
Epi4K Consortium (2016) De novo mutations in SLC1A2 and CACNA1A are important causes of epileptic encephalopathies. Am J Hum Genet 99(2):287–298
Tukiainen R, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A et al (2017) Landscape of X chromosome inactivation across human tissues. Nature 550(7675):244–248
Peeters SB, Cotton AM, Brown CJ (2014) Variable escape from X-chromosome inactivation: identifying factors that tip the scales towards expression. Bioessays 36(8):746–756
Cotton AM, Price EM, Jones MJ, Balaton BO, Kobor MS, Brown CJ (2015) Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum Mol Genet 24(6):1528–1539
Toksoy G, Durmus H, Aghayev A, Bagirova G, Sevinc Rustemoglu B, Basaran S, Avci S, Karaman B, Parman Y, Altunoglu U, Yapici Z, Tekturk P, Deymeer F, Topaloglu H, Kayserili H, Oflazer-Serdaroglu P, Uyguner ZO (2019) Mutation spectrum of 260 dystrophinopathy patients from Turkey and important highlights for genetic counseling. Neuromuscul Disord 29(8):601–613
Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM (2005) Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CPG methylation during early development. Dev Cell 8(1):31–42
Dandulakis MG, Meganathan K, Kroll KL, Bonni A, Constantino JN (2016) Complexities of X chromosome inactivation status in female human induced pluripotent stem cells-a brief review and scientific update for autism research. J Neurodev Disord 8:22. https://doi.org/10.1186/s11689-016-9155-8
Geens M, Chuva De Sousa Lopes SM (2017) X chromosome inactivation in human pluripotent stem cells as a model for human development: back to the drawing board? Hum Reprod Update 23(5):502–532
Bragança J, Lopes JA, Mendes-Silva L, Almeida Santos JM (2019) Induced pluripotent stem cells, a giant leap for mankind therapeutic applications. World J Stem Cells 11(7):421–430
Hymes J, Fleischhauer K, Wolf B (1995) Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem Mol Med 56(1):76–83
Küry S, Ramaekers V, Bézieau S, Wolf B (2016) Clinical utility gene card for: Biotinidase deficiency-update 2015. Eur J Hum Genet 24(7):3–5. https://doi.org/10.1038/ejhg.2015.246
Acknowledgements
Core Facility Bioinformatics of CEITEC Masaryk University is gratefully acknowledged for obtaining of the scientific data presented in this paper. Computational resources were provided by the CESNET LM2015042 and the CERIT Scientific Cloud LM2015085, provided under the programme “Projects of Large Research, Development, and Innovations Infrastructures”. The authors are thankful to the family for its participation, and all physicians and medical staff providing the specialized medical examinations supporting and completing the diagnosis of our patients. This work greatly extends the poster presentation by the second author MR on AES 2019 Annual Meeting.
Funding
This work was supported by the grant of the Ministry of Health, Czech Republic, Conceptual Development of Research Organization (FNBr, 65269705); by the grant of the Faculty of Science, Masaryk University, Brno, Czech Republic (MUNI/A/1127/2019); and by the grant of the Ministry of Health, Czech Republic, Czech Health Research Council (NU20-07-00145).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. MW analyzed and interpreted the NGS data regarding the probands’ phenotypes using relevant literature, bioinformatics databases, and in silico tools. MW was a major contributor in writing the manuscript. MR provided specialized neurological examination and clinical data from the Clinic of Children’s Neurology. JO processed raw NGS data using advanced bioinformatics tools through a unique multistep pipeline. VV, EH, and HP participated in microarray and NGS data analyses and targeted Sanger sequencing analyses. LK performed X chromosome inactivation analysis. MV performed the cytogenetic analysis of karyotype. RG provided specialized genetic counselling for the probands and their family and interpreted the findings in the clinical context. PK contributed towards the interpretation of data and performed general scientific supervision and general critical revision of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All procedures performed involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Approval was obtained from the Ethics Committee of the University Hospital Brno.
Informed Consent
Written institutional informed consent (University Hospital Brno, Czech Republic) was obtained from the parents of the patients before the procedure of genetic analyses. Our case report does not include any personal information leading to the identification of any participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wayhelova, M., Ryzí, M., Oppelt, J. et al. Novel familial IQSEC2 pathogenic sequence variant associated with neurodevelopmental disorders and epilepsy. Neurogenetics 21, 269–278 (2020). https://doi.org/10.1007/s10048-020-00616-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-020-00616-3