Abstract
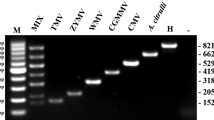
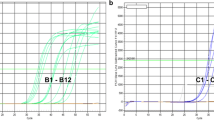
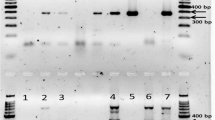
The complete sequence of the cucumber mosaic virus (CMV) satellite RNA (satRNA) gene that was controlled by the cauliflower mosaic virus (CaMV) 35S promoter (P-35S) and the Agrobacterium nopaline synthase terminator (T-nos) was first identified in an unapproved genetically modified (GM) tomato (Solanum lycopersicum L.), and a duplex polymerase chain reaction (PCR) method was developed based on the CMV satRNA nucleotide sequence. To detect the unapproved GM tomato, the metallocarboxypeptidase inhibitor (Mcpi) gene was selected as an endogenous reference gene for tomato and was validated using 13 different crops. The primer pair PTD-F/R was designed to amplify the junction sequences between the 35S promoter and CMV satRNA gene introduced in the unapproved GM tomato, and its specificity was also validated using several different GM events. The detection limit of the duplex PCR method is approximately 1 copy. Using the duplex PCR method, 35 processed tomato foods and 13 tomato seeds were analyzed. Of these samples, 2 GM tomato seeds were identified using the duplex PCR method. The results indicate that this duplex PCR method could be useful for detecting the unapproved GM tomato.
Similar content being viewed by others
References
MIFAFF. Statistics on Production of Greenhouse Vegetable and Greenhouse Facilities for Vegetable 2009. Ministry for Food, Agriculture, Forestry, and Fisheries, Gwacheon, Korea. p. 9 (2010)
Korea Agricultural Trade Information (KATI), Seoul, Korea, Trade Statistics. Available from: http://www.kati.net. Accessed Dec. 3, 2011.
International Service for the Acquisition of Agri-biotech Applications (ISAAA), Ithaca, NY, USA, GM Approval Database. Available from: http://www.isaaa.org. Accessed Dec. 3, 2011.
Fischhoff DA, Bowdish K, Perlak F, Marrone PG, McCormick SM, Niedermeyer JG, Dean DA, Kusano-Kretzmer K, Mayer EJ, Rochester DE, Rogers SG, Fraley RT. Insect tolerant transgenic tomato plants. Biotechnol. Adv. 5: 807–813 (1987)
Yang L, Huifeng S, Aihu P, Jianxiu C, Cheng H, Dabing Z. Screening and construct-specific detection methods of transgenic Huafan No 1 tomato by conventional and real-time PCR. J. Sci. Food Agr. 85: 2159–2166 (2005)
Ouyang B, Chen YH, Li HX, Qian CJ, Huang SL, Ye ZB. Transformation of tomatoes with osmotin and chitinase genes and their resistance to Fusarium wilt. J. Hortic. Sci. Biotech. 80: 517–522 (2005)
Randhawa GJ, Singh M, Chhabra R, Guleria S, Sharma R. Molecular diagnosis of transgenic tomato with osmotin gene using multiplex polymerase chain reaction. Curr. Sci. India 96: 689–694 (2009)
Huang J, Rozelle S. China aggressively pursuing horticulture and plant biotechnology. Calif. Agr. 58: 112–113 (2004)
Palukaitis P, Roossinck MJ, Dietzgen RG, Francki RI. Cucumber mosaic virus. Adv. Virus Res. 41: 281–348 (1992)
Lin CH, Sheu F, Lin HT, Pan TM. Allergenicity assessment of genetically modified cucumber mosaic virus (CMV) resistant tomato (Solanum lycopersicon). J. Agr. Food Chem. 58: 2302–2306 (2010)
Morroni M, Thompson JR, Tepfer M. Twenty years of transgenic plants resistant to cucumber mosaic virus. Mol. Plant Microbe Int. 21: 675–684 (2008)
RIC. China Tomato Products Industry Report, 2009–2010. Res. In China, Beijing, China. pp. 1–11 (2010)
KFDA. Amendment to The Food Standard & Specifications Notification No. 2007-68. Korea Food & Drug Administration, Osong, Korea (2007)
Crescenzi A, Barbarossa L, Cillo F, Di Franco A, Vovlas N, Gallitelli D. Role of cucumber mosaic virus and its satellite RNA in the etiology of tomato fruit necrosis in Italy. Arch. Virol. 131: 321–333 (1993)
Saito Y, Komari T, Masuta C, Hayashi Y, Kumashiro T, Takanami Y. Cucumber mosaic virus-tolerant transgenic tomato plants expressing a satellite RNA. Theor. Appl. Genet. 83: 679–683 (1992)
Harrison BD, Mayo MA, Baulcombe DC. Virus resistance in transgenic plants that express cucumber mosaic virus satellite RNA. Nature 328: 799–802 (1987)
Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual (3 volume set). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA. pp. 14–19 (2001)
Kim JH, Kim SY, Lee HJ, Kim YR, Kim HY. An event-specific DNA microarray to identify genetically modified organisms (GMOs) in processed foods. J. Agr. Food Chem. 58: 6018–6026 (2010)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, JH., Lee, JE. & Kim, HY. Duplex polymerase chain reaction method for detection of unapproved genetically modified tomato (Solanum lycopersicon L.) with cucumber mosaic virus (CMV) satellite RNA gene. Food Sci Biotechnol 21, 823–827 (2012). https://doi.org/10.1007/s10068-012-0106-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-012-0106-1