Abstract
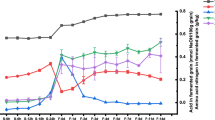
The Ginkgo epicarp is usually discarded due to contained butyric acid causing an unpleasant odor. Control of butyric acid in the Ginkgo epicarp was achieved by fermenting with doenjang under different fermentation conditions. The contents of butyric acid decreased 3.2× when Ginkgo epicarp was fermented at 30°C for 6 months. Microbes in doenjang were probably responsible for the reduction in the butyric acid content. Microbial communities were analyzed during fermentation. Communities were isolated using selective media and analyzed using metagenomic pyrosequencing based on 16S rRNA sequences. Changes in the microbial communities were confirmed at the species level. Staphylococcus gallinarum was the dominant species and maintained a constant level during fermentation. The possibility of a new fermented food with Ginkgo epicarp is suggested.
Similar content being viewed by others
References
Itil TM, Eralp E, Ahmed I, Kunitz A, Itil KZ. The pharmacological effects of Ginkgo biloba, a plant extract, on the brain of dementia patients in comparison with tacrine. Psychopharmacol. Bull. 34: 391–397 (1998)
Major RT. The Ginkgo, the most ancient living tree to the existence of Ginkgo biloba L. to pests accounts in part for the longevity of this species. Science 157: 1270–1273 (1967)
Singh B, Karu Pyshpinder, Gopichand, Singh RD, Ahuja PS. Biology and chemistry of Ginkgo biloba. Fitoterapia 79: 401–418 (2008)
Wu XY, Mao G, Zhao T, Zhao J, Li F, Liang L, Yang L. Isolation, purification, and in vitro anti-tumor activity of polysaccharide from Ginkgo biloba sarcotesta. Carbohyd. Polym. 86: 1073–1076 (2011)
Chen SX, Wu L, Yang XM, Jiang XG, Li LG, Zhang RX. Comparative molluscicidal action of extract of Ginko biloba sarcotesta, arecoline and niclosamide on snail hosts of Schistosoma japonicum. Pestic. Biochem. Physiol. 89: 237–241 (2007)
Sasaki K, Wada K, Haga M. Chemistry and biological activities of Ginkgo biloba. Stud. Nat. Prod. Chem. 28: 165–198 (2003)
Pan W, Luo P, Fu R, Gao P, Long Z, Xu F, Xiao H, Liu S. Acaricidla activity against Panonychus citri of a Ginkgo biloba. Pest Manag. Sci. 62: 283–287 (2006)
Mahadevan S, Park Y. Multifaceted Therapeutic benefits of biloba L.: Chemistry, efficacy, safety, and uses. J. Food Sci. 73: R14–R19 (2008)
Guyot C, Bonnafont C, Lesschaeve I, Issanchou S, Boilley A, Spinnler HE. Effect of fat content on odor intensity of three aroma compounds in model emulsions: δ-Decalacone, diacetyl, and butyric acid. J. Agr. Food Chem. 44: 2341–2348 (1996)
Shin ZI, Yu R, Park SA, Chung DK, Ahn CW, Nam HS, Kim KS, Lee HJ. His-His-Leu, an angiotensin I converting enzyme inhibitory peptide derived from Korean soybean paste, exerts antihypertensive activity in vivo. J. Agr. Food Chem. 49: 3004–3009 (2001)
Bernard FG, Alexandre Z, Robert M, Catherine M. Production and characterization of bioactive peptides from soy hydrolysate and soyfermented food. Food Res. Int. 37: 123–131 (2004)
Cho DH, Lee WH. Microbiological studies of Korean native soy sauce fermentation: A study on the microbora of fermented Korean Maeju loaves. J. Korean Agric. Chem. Soc. 13: 35–42 (1970)
Lee JS, Kwon SJ, Chung SW, Choi YJ, Yoo JY, Chun DH. Changes of microorganisms, enzyme activities, and major components during the fermentation of Korean traditional Doenjang and Kochujang. Korean J. Microbiol. Biotechnol. 24: 247–253 (1996)
Yoo SK, Cho WH, Kang SM, Lee SH. Isolation and identification of microorganisms in Korean traditional soybean paste and soybean sauce. Korean J. Appl. Microbiol. Biotechnol. 27: 113–117 (1999)
Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 35: e130 (2007)
Kim YS, Kim MC, Kwon SW, Kim SJ, Park IC, Ka JO, Weon HY. Analysis of bacterial communities in Meju, a Korean traditional fermented soybean bricks, by cultivation-based and pyrosequencing methods. J. Microbiol. 49: 340–348 (2011)
Roh SW, Kim KH, Nam YD, Chang HW, Park EJ, Bae JW. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J. 4: 1–16 (2010)
Al-Zoreky N, Sandine WE. A selective and differential agar medium. J. Food Sci. 56: 1729 (1991)
Lee MK, Park WS, Kang KH. Selective media for isolation and enumeration of lactic acid bacteria from kimchi. J. Korean Soc. Food Sci. Nutr. 25: 754–760 (1996)
Weisburg WG, Bams SM, Pellertier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173: 697–703 (1991)
Soumitesh C, Danica H, Michele B, Nancy C, David A. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69: 330–339 (2007)
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200 (2011)
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Sys. Evol. Microbiol. 57: 2259–2261 (2007)
Needleman SB, Wunsch CD. A general method applicable to the sarch for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48: 443–453 (1970)
Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659 (2006)
Chao A. Nonparametric estimation of the number of classes in a population. Scand. J. Statist. 11: 265–270 (1984)
Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika 40: 237–264 (1953)
Kenneth LH, Gerald VB, Daniel S. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56: 1459–1461 (1975)
Schloss PD, Westcott SL. Ryabin T, Hall JR, Hartmann M. Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahi JW, Stres B, Thallinger GG, Van Harn DJ, Weber CF. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75: 7537–7541 (2009)
Jacob TJC, Fraser C, Wang L, Walker V, O’Connor S. Psychophysical evaluation of responses to pleasant and mal-odour stimulation in human subjects; adaptation, dose response and gender differences. Int. J. Psychophysiol. 48: 67–80 (2003)
Kim YS, Jeong DY, Hwang YT, Uhm TB. Bacterial community profiling during the manufacturing process of traditional soybean paste by pyrosequencing method. Korean J. Microbiol. 47: 275–280 (2011)
Hill TC, Walsh KA, Harris JA, Moffett BF. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43: 1–11 (2003)
Kim TW, Lee JH, Park MH, Kim HY. Analysis of bacterial and fungal communities in Janpanese and Chines-fermented soybean pastes using nested PCR-DGGE. Curr. Microbiol. 60: 315–320 (2010)
Kempf M, Theobald U, Fiedler HP. Production of the antibiotic gallidermin by Staphylococcus gallinarum-development of a scale-up procedure. Biotech. Lett. 22: 123–128 (2000)
Drosinos EH. Paramithiotis S, Kolovos G, Tsikouras I, Metaxopoulos I. Phenotypic and technological diversity of lactic acid bacteria and staphylococci isolated from traditionally fermented sausages in Southern Greece. Food Microbiol. 24: 260–270 (2007)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Park, S., Song, KM., Kim, HJ. et al. Characterization and use of microbial communities in Doenjang to control the unpleasant odor of Ginkgo epicarp. Food Sci Biotechnol 23, 1959–1964 (2014). https://doi.org/10.1007/s10068-014-0267-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-014-0267-1