Abstract
Background
Accumulating evidence indicates that long non-coding RNAs (lncRNAs) participate in progression of gastric cancer (GC). Nevertheless, the function and expression level of DLX6-AS1 in GC remain unknown.
Methods
We explored the sequencing data of DLX6-AS1 downloaded from The Cancer Genome Atlas. The expression of DLX6-AS1, miR-204-5p and OCT1 in 56 GC patients and GC cell lines was quantified by qRT-PCR and western blotting. Furthermore, we performed in vitro functional assays to assess proliferation, invasion and migration of GC cells by knockdown of DLX6-AS1. The expression level of epithelial–mesenchymal transition (EMT)-related genes was also determined by qRT-PCR and western blotting. Actin remodeling was detected by F-actin phalloidin staining. The luciferase reporter assay and chromatin immunoprecipitation assay was utilized to confirm the bioinformatic prediction. The function of the DLX6-AS1/miR-204-5p/OCT1 axis in GC proliferation was clarified by rescue assays.
Results
We first demonstrated that DLX6-AS1 was upregulated in GC tissues and cell lines and was associated with T3/T4 invasion, distant metastasis and poor clinical prognosis. Further functional analysis showed that downregulation of DLX6-AS1 inhibited GC cell proliferation, migration, invasion and EMT in vitro. Mechanistic investigation indicated that DLX6-AS1 acted as a cancer-promoting competing endogenous RNA (ceRNA) by binding miR-204-5p and upregulating OCT1. Moreover, the transcription factor OCT1 was confirmed to enhance DLX6-AS1 expression by targeting the promoter region.
Conclusions
This study revealed that OCT1-induced DLX6-AS1 promoted GC progression and the EMT via the miR-204-5p/OCT1 axis, suggesting that this lncRNA might be a promising prognostic biomarker and therapeutic target for GC.
Similar content being viewed by others
Introduction
According to global cancer statistics 2018 [1], there were approximately 1.03 million new cases and 0.78 million deaths from gastric cancer (GC) in that year. The GC incidence and mortality were ranked third and fifth, respectively, worldwide. At present, surgery combined with radiotherapy and chemotherapy is the primary curative treatment for the disease. However, GC is usually diagnosed in the progressing stage due to its atypical symptoms. Hence, there is an urgent need to improve the early diagnosis and improve the prognosis of GC patients by identifying novel targets.
The Encyclopedia of DNA Elements project has demonstrated that approximately 98% of the genome does not encode proteins and is thus regarded as non-coding RNA (ncRNA) [2]. As a type of ncRNA, long non-coding RNAs (lncRNAs) are defined as a class of transcripts of more than 200 nucleotides without an open reading frame [3]. Accumulating studies have reported that lncRNAs participate in diverse neoplastic biological processes, including carcinogenesis, proliferation, metastasis and the epithelial-mesenchymal transition (EMT). For this reason, researches into lncRNAs have attracted a great deal of attention. Interestingly, lncRNAs may play opposing roles in different organ tumors. For instance, the lncRNA FER1L4 is overexpressed in glioblastoma and would be regarded as a poor diagnostic biomarker [4]. However, FER1L4 exerts a tumor-suppressive effect on hepatocellular cancer [5].
Recent researches have revealed that lncRNAs act as competing endogenous RNAs (ceRNAs) by binding microRNAs (miRNAs) with shared response elements and dysregulating tumor oncogenes and suppressor genes [6, 7]. Studies have indicated that lncRNAs regulate EMT-related proteins by binding miRNAs and are thus recognized as a promising therapeutic target [8, 9]. For instance, lncRNA MALAT1 acts as a miR-204-5p sponge and upregulates expression of the EMT-related marker SLUG in lung adenocarcinoma [10], while lncRNA UCA1 targets miR-204-5p and upregulates ZEB1 expression in glioma [11].
By analyzing the cancer genome atlas (TCGA) database, we identified a survival-related lncRNA, DLX6-AS1, which was upregulated in GC. The lncRNA DLX6-AS1 is a 1990 bp non-coding transcript located at chromosomal band 7q21.3 that has been reported to be overexpressed in lung adenocarcinoma [12], hepatocellular cancer [13, 14], renal cell cancer [15], osteosarcoma [16], glioma [17] and pancreatic cancer [18]. Nevertheless, the carcinogenesis and regulatory mechanism of DLX6-AS1 in GC remains unknown. In the present study, we first verified that DLX6-AS1 was overexpressed in GC. We then explored the oncogenic function of DLX6-AS1 in GC development by loss-of-function analysis. Through bioinformatics prediction and a series of experiments, we hypothesized and verified that DLX6-AS1 could promote GC cell proliferation, migration and invasion by directly targeting the miR-204-5p and upregulating OCT1 (POU2F1). Furthermore, we found that transcription factor OCT1 could activate the expression of DLX6-AS1 by binding to the promoter region. In addition, we performed rescue assays to clarify the role of the DLX6-AS1/miR-204-5p/OCT1 positive feedback loop in GC.
Materials and methods
Sample collection from the database
We downloaded all of the GC transcriptome profiles and clinical data from TCGA database. Up to September 2018, the public database included the expression profiles of 375 GC tissues and 32 normal tissues obtained by RNA-seq (level 3). Our research conformed to the guidelines published in TCGA. We took the logarithm of the expression difference between normal and tumor samples.
Patients and tissues
From September 2017 to July 2018, we collected a total of 56 paired GC tissue and adjacent noncancerous tissue (> 5 cm from cancer tissue) samples, as well as clinical data, from the GC patients who underwent stomach resection at the Fourth Affiliated Hospital (Cancer Institute) of China Medical University. The patients had not received any previous therapy before surgery. The enrolled patients provided signed informed consent. The TNM stage histological grade was confirmed based on the 8th American Joint Committee on Cancer (AJCC) system. This study was approved by the Clinical Research Ethics Committee of China Medical University and was performed according to the guidelines of the Declaration of Helsinki.
Cell lines and culture
The seven GC cell lines including MKN-7, MKN-28, MGC-803, HGC-27, MKN-45, AGS and SGC-7901 along with the normal intestinal cell line GES-1 and human embryonic kidney (HEK) cell line 293T were purchased from the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI 1640 medium (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA). Moreover, 100 U/mL penicillin, and 100 μg/mL streptomycin were added to the medium to prevent infection. All cells were cultured at 37 °C in a humidified incubator with 5% CO2.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted using RNAios Plus (Takara, Otsu, Shiga, Japan). The quality and concentration of the isolated RNA were analyzed using a Nano drop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA (800 ng) was converted to cDNA using the PrimeScript™ RT reagent Kit (Takara) with gDNA Eraser. To analyze the expression of miRNA, we performed reverse transcription with the miR-100-3p, miR-195-3p, miR-145-5p and miR-204-5p specific stem-loop primers and Hairpin-it™ miRNA Quantitation Kit (GenePharma, Suzhou, Jiangsu, China). We utilized a LightCycler 96 (Roche Diagnostic, Basel, Switzerland) to perform qRT-PCR using SYBR Green (Takara). The conditions of the two-step amplification reaction were as follows: pre-incubation at 95 °C for 30 s, then 40 cycles of 95 °C for 10 s and 60 °C for 30 s. GAPDH and U6 expression was utilized as the endogenous control for mRNA/lncRNA and miRNA, respectively. Expression level analysis was performed in accordance with the \(2^{{ - \Delta \Delta C_{T} }}\) method [19]. Specific primers for RNAs are shown in Supplementary Table 1.
Transfection
According to the manufacturers’ protocol, all the small interfering RNAs (siRNAs) (GenePharma) including siRNA-DLX6-AS1 (#1, #2, #3) and siRNA-OCT1 (#1, #2, #3) against different sites in DLX6-AS1 and OCT1, and negative control siRNA (si-NC), were transfected into SGC7901 and AGS cells using the Lipofectamine 3000 Reagent (Invitrogen). MiR-204-5p inhibitor as well as miRNA negative control (miR-NC) purchased from GenePharma were transfected into cells by the same method. The sequences of siRNAs and miRNA inhibitor are shown in Supplementary Table 2. The silencing effect was confirmed by qRT-PCR or western blotting.
Cell proliferation assay
The cell counting kit-8 (CCK-8) assay was performed to evaluate cell proliferation capacity. A total of 3000 transfected cells were seeded into each of five replicate wells, then 10 μL CCK-8 solution (Solarbio, Beijing, China) was added into each well. After incubation for 1 h, the absorbance at 450 nm was read on a microplate reader to measure the cell viability every 24 h for 4 days.
Colony formation assay
Approximately 200 transfected cells were plated into each well of 6-well plates. After 2 weeks of incubation, we were able to observe colonies with the naked eye. The colonies were then fixed using 4% paraformaldehyde for 15 min and stained using 0.1% Crystal Violet (Solarbio) for 10 min. Formed colonies were counted and photographed.
Transwell assay
After transfection of SGC-7901 and AGS cells for 24 h, the cells were collected and cell density was adjusted to 1 × 105/mL with FBS-free RPMI 1640 medium. Approximately 20,000 cells were plated into the upper insert of a Transwell chamber (Corning, New York, NY, USA) with or without Matrigel™ (Corning) to evaluate invasion or migration functions. Medium with 20% FBS was then added into the lower chamber as the stimulus. After 24 h (migration) and 48 h (invasion), the chambers were fixed and stained as described above. To eliminate the effect of different proliferation abilities on the results, we further performed Transwell assays of transfected SGC-7901 and AGS cells at progressive concentrations of chemoattractant (0, 5, 10, 20, and 40%) under si-DLX6-AS1 treatment. The cells that migrated to the bottom of the filter were counted in 10 random fields from random portions of the filter.
Western blot analysis
Cells were collected and lysed in RIPA lysis buffer (Solarbio) on ice, then extracted proteins were quantified and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Solarbio). The PVDF membranes were blocked using 5% skimmed milk for 2 h with gentle shaking, then incubated with primary antibodies diluted with primary antibody dilution buffer (Solarbio) as follows: E-cadherin (1:100; Abcam, Cambridge, MA, USA), N-cadherin (1:2000; Abcam), SLUG (1:2000; Abcam), MMP9 (1:2000; Abcam), OCT1 (1:1500; Abcam) and β-actin (1:2500; Abcam). After incubation at 4 °C overnight, the membranes were thoroughly washed and incubated with horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit, 1:5000; Wanleibio, Shenyang, Liaoning, China) at room temperature for 2 h. We utilized a chemiluminescence western blotting detection system (Tanon, Shanghai, China) to analyze the protein bands.
F-actin phalloidin staining and fluorescence microscopy
The slides of transfected cells were fixed using 4% paraformaldehyde for 10 min and permeabilized with 0.5% Triton X-100 (Solarbio) for 5 min. Each slide was incubated with 200 μL FITC-labeled phalloidin (100 nM, Solarbio) containing 1% bovine serum albumin at 37 °C for 1 h in the dark. Then, the nuclei were stained with 200 μL DAPI solution (100 nM, Solarbio). We utilized confocal microscopy (Nikon, Tokyo, Japan) to image transfected cells.
Bioinformatics prediction and luciferase reporter assay
We searched the possible miRNAs that may be involved in crosstalk with DLX6-AS1 by applying two different target prediction algorithms miRcode (https://www.mircode.org/) [20] and DIANA-LncBase (https://www.microrna.gr/LncBase) [21]. After identifying miR-204-5p as the research target, we identified and predicted the most likely miR-204-5p binding sites in the DLX6-AS1 3′-UTR regions. PmirGLO (Promega, Madison, WI, USA) was used to construct the luciferase reporter vectors DLX6-AS1-wild type (WT) and DLX6-AS1-Mutant (MUT). A QuickMutation™ Site-Directed Mutagenesis Kit (Beyotime, Nantong, Jiangsu, China) was utilized to introduce the mutations of the miR-204-5p potential binding sites. HEK293T, SGC-7901 and AGS cell lines were seeded into 24-well plates and co-transfected with DLX6-AS1-WT or MUT vectors and miR-204-5p mimic or miR-NC. The interaction between miR-204-5p and the 3′-UTR regions of OCT1 was predicted by miRTarBase (https://miRTarBase.mbc.nctu.edu.tw/) [22] and verified using the same method. The miRTarBase data of miR-204-5p are shown in Supplementary Fig. 1. Moreover, we identified possible binding sites between OCT1 and the DLX6-AS1 promotor using JASPAR (https://jaspar.genereg.net) [23]. The two predicted binding sites and the corresponding mutant sequences (MUT1 and MUT2) were introduced into the pGL3 plasmid vector (Promega). SGC-7901 and AGS cell lines were seeded into 24-well plates and co-transfected with the vectors described above and the OCT1 overexpression vector or normal control (NC) groups. A luciferase reporter assay kit (Promega) was used to evaluate the luciferase activity according to the manufacturers’ instructions.
Chromatin immunoprecipitation (ChIP) assay
We performed a ChIP assay using a Magna ChIP Kit (Millipore, Bedford, MA, USA). The top five possible sequences between OCT1 and the DLX6-AS1 promoter were obtained from JASPAR. SGC-7901 and AGS cell lines were cross-linked by adding formaldehyde and terminated by adding glycine. The 200–1000 bp fragments were generated from lysates through sonication. Specific DNA–protein complexes were immunoprecipitated using an incubating antibody (anti-OCT1 and lgG) (Millipore). Purified DNA was quantified and analyzed by qRT-PCR. The IgG group and the input group were used as the negative control group and the positive control group, respectively.
Statistical analysis
All statistical analyses were performed and visualized using SPSS V18.0 (SPSS, Chicago, IL, USA) and Prism 7.0 (GraphPad, La Jolla, CA, USA) software. Student’s t test or the chi-squared test were used for two-group comparisons and multiple group comparisons, as appropriate. The correlation between lncRNA, miRNA and mRNA were analyzed by Spearman’s correlation analysis. Kaplan–Meier curve analysis was performed to evaluate the overall survival. All data are expressed as the mean ± standard deviation (SD) and all experiments were repeated three times. Differences with a value of P < 0.05 or < 0.01 were regarded as statistically significant.
Results
DLX6-AS1 is upregulated in GC
We collected and analyzed the RNA-seq data of 375 GCs and 32 normal samples from TCGA database. The results showed that DLX6-AS1 was overexpressed in GC tissues (P = 0.0004, Fig. 1a). After excluding samples that lacked data regarding survival time and status, a total of 372 GC patients were divided into two groups for Kaplan–Meier curve analysis according to the median value of DLX6-AS1 expression. The result indicated that high expression of DLX6-AS1 was significantly associated with poor prognosis of GC patients (P = 0.042, Fig. 1b). Furthermore, we performed qRT-PCR to analyze the expression level of DLX6-AS1 in 56 GC and corresponding noncancerous tissues. The results of qRT-PCR were consistent with the data in TCGA database (P < 0.01, Fig. 1c). Then, we divided the GC patients into an above-median expression group and a below-median expression group. The expression level in patients was associated with T3/T4 invasion (P = 0.035) and distant metastasis (P = 0.018), while DLX6-AS1 expression was not associated with the other clinicopathological characteristics (Supplementary Table 3). This suggested that DLX6-AS1 may have some effect on tumor progression.
DLX6-AS1 is upregulated in GC tissues and cells. a Relative expression of DLX6-AS1 in GC and normal tissues from TCGA database. b Kaplan–Meier curve analysis of the correlation of DLX6-AS1 expression and overall survival from TCGA database. c qRT-PCR analysis of DLX6-AS1 expression in 56 GC tissues and corresponding adjacent noncancerous tissues. Data are shown as − ΔΔCT values. d qRT-PCR analysis of DLX6-AS1 expression in a normal gastric cell line (GES-1) and seven GC cell lines (MKN-7, MKN-28, SGC-7901, MCG-803, AGS, MKN-45 and HGC-27). Data are presented as the mean ± SD. *P < 0.05, **P < 0.01
To elucidate the function of DLX6-AS1, we performed qRT-PCR to analyze the expression of DLX6-AS1 in cell lines. As shown in Fig. 1d, DLX6-AS1 was upregulated in SGC-7901, MCG-803, AGS, MKN-45, HGC-27, MKN-7 and MKN-28 compared to GES-1 (P < 0.01). As SGC-7901 and AGS expressed the highest levels of DLX6-AS1, we chose these two cell lines for functional and mechanistic studies.
DLX6-AS1 promotes GC proliferation, migration, invasion, and EMT-related gene expression in vitro
To elucidate the function of DLX6-AS1, DLX6-AS1 was knocked down by transfecting three different siRNAs into cells. At 48 h after transfection, we performed qRT-PCR to verify that DLX6-AS1 was effectively suppressed by siRNA #2 and #3 (P < 0.01, Fig. 2a). The results of CCK-8 assay revealed that DLX6-AS1 downregulation reduced cell growth ability (P < 0.01, Fig. 2b). Similarly, the number of visible colonies was decreased in cells transfected with DLX6-AS1 knockdown compared to transfection with si-NC (P < 0.01, Supplementary Fig. 2a). The influence of DLX6-AS1 on invasion and migration was investigated by Transwell assay. We observed that siRNA transfection significantly inhibited the migration and invasion capacity of GC cells (P < 0.01, Fig. 2c). Furthermore, we obtained similar results at different concentrations of chemoattractant (P < 0.01, Supplementary Figs. 3, 4).
DLX6-AS1 promotes proliferation, invasion, migration and EMT-related gene expression of GC cells in vitro. a The qRT-PCR analysis of DLX6-AS1 in SGC-7901 and AGS cells transfected with si-NC or si-DLX6-AS1. b CCK-8 assays were performed to evaluate the growth ability of SGC-7901 and AGS cells transfected with si-NC or si-DLX6-AS1. c Transwell assays were performed to assess the invasion or migration function of SGC-7901 and AGS cells transfected with si-NC or si-DLX6-AS1. d, e The mRNA and protein levels of EMT-related genes in SGC-7901 and AGS cell lines treated with si-NC or si-DLX6-AS1 were determined by qRT-PCR and western blotting. f F-actin staining assays were used to detect the actin cytoskeleton changes in SGC-7901 and AGS cells transfected with si-NC or si-DLX6-AS1. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01
To further investigate the mechanism by which DLX6-AS1 promotes proliferation, invasion and metastasis of GC, we performed qRT-PCR and western blotting to measure the epithelial marker E-cadherin, the mesenchymal markers N-cadherin and SLUG, and the invasion marker MMP9 at transcriptional and translational levels in SGC-7901 and AGS cell lines. As shown in Fig. 2d, e knockdown of DLX6-AS1 inhibited N-cadherin (P < 0.05), MMP9 (P < 0.05), and SLUG (P < 0.05) expression and promoted E-cadherin (P < 0.01) expression according to qRT-PCR and western blot results. Furthermore, we observed that transfected cells exhibited polygonal, rounded and epithelial-like morphology under si-DLX6-AS1 treatment (Supplementary Fig. 2b). Because actin remodeling has been verified as a critical mechanism in the process of the EMT [24], we then performed phalloidin staining to reveal the effect of DLX6-AS1 downregulation on F-actin organization. As shown in Fig. 2f, the distribution of both the actin cytoskeleton and filopodia were significantly decreased when DLX6-AS1 was knocked down. In summary, DLX6-AS1 participated simultaneously in GC progression and EMT.
DLX6-AS1 acts as a ceRNA by directly binding miR-204-5p
To explore the possible molecular mechanism of DLX6-AS1 in GC, we performed bioinformatics prediction to identify the target of DLX6-AS1 by intersecting the predicted results between miRcode, DIANA-LncBase and downregulated miRNAs in TCGA. As shown in Fig. 3a, we identified four miRNAs (miR-100-3p, miR-145-5p, miR-195-3p, and miR-204-5p) that showed the possibility of interacting with DLX6-AS1. Then, we performed qRT-PCR to detect expression of the corresponding miRNAs. The results showed that only miR-204-5p was upregulated in si-DLX6-AS1-transfected cell lines (P < 0.01, Fig. 3b). Consequently, we selected miR-204-5p for further study. The potential base pair binding sites between miR-204-5p and DLX6-AS1 are shown in Fig. 3c. To further verify our hypothesis, we co-transfected luciferase reporter vectors and miR-204-5p mimic or miR-NC into the HEK293T, SGC-7901 and AGS cell lines. As shown in Fig. 3d, luciferase activity was significantly reduced in the DLX6-AS1-WT together with miR-204-5p mimic reaction system (P < 0.05). This indicated that miR-204-5p interacted with DLX6-AS1 through sequence matching.
DLX6-AS1 acts as a ceRNA by directly binding miR-204-5p. a The target miRNAs of DLX6-AS1 were predicted using three online databases (TCGA, miRcode, and DIANA-LncBase). b The qRT-PCR analysis of miR-100-3p, miR-145-5p, miR-195-3p and miR-204-5p in SGC-7901 and AGS cells transfected with si-NC or si-DLX6-AS1. c Binding sites between miR-204-5p and DLX6-AS1 were predicted using the bioinformatics database. d DLX6-AS1-WT vectors or DLX6-AS1-MUT vectors and miR-204-5p mimics and miR-NC were transfected into HEK293T, SGC-7901 and AGS cells, and the luciferase activity was measured. e The qRT-PCR analysis of miR-204-5p expression in 56 GC tissues and corresponding adjacent noncancerous tissues. Data are shown as − ΔΔCT values. f The qRT-PCR analysis of miR-204-5p expression in a normal gastric cell line (GES-1) and five GC cell lines (SGC-7901, MCG-803, AGS, MKN-45 and HGC-27). g The correlation between DLX6-AS1 expression and miR-204-5p expression was measured in 56 GC patients by Spearman’s correlation analysis. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01
We performed qRT-PCR to analyze the expression level of miR-204-5p. As shown in Fig. 3e, f miR-204-5p was obviously reduced in GC cell lines and tumor tissues (P < 0.01), which is in accordance with previous findings. Furthermore, miR-204-5p expression was negatively correlated with tumor size (P = 0.003), invasion (P = 0.035), distant metastasis (P < 0.001) and AJCC staging (P = 0.033, Supplementary Table 3). Spearman’s correlation analysis indicated a negative relationship between DLX6-AS1 and miR-204-5p (P = 3.94E−9, r = − 0.628, Fig. 3g). Hence, the above results revealed that DLX6-AS1 could act as a ceRNA by binding miR-204-5p.
Tumor promotor effects of DLX6-AS1 are partially mediated by miR-204-5p
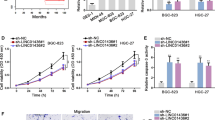
To further confirm the role of miR-204-5p in the effect of DLX6-AS1 on GC, SGC-7901 and AGS cell lines were co-transfected with si-DLX6-AS1 #3, miR-204-5p inhibitor, si-NC and miR-NC for functional assays (Fig. 4a). The results of CCK8, colony formation and Transwell assays indicated that miR-204-5p inhibitor promoted GC progression (P < 0.01). Moreover, miR-204-5p inhibitor partially rescued the antitumor function of si-DLX6-AS1 #3 (Fig. 4b, c, Supplementary Fig. 5a). These phenomena confirmed that miR-204-5p played a critical role in the oncogenic function of DLX6-AS1.
Tumor promotor effects of DLX6-AS1 mediated by miR-204-5p. a The qRT-PCR analysis of miR-204-5p expression in SGC-7901 and AGS cells co-transfected with si-NC or si-DLX6-AS1 #3 and miR-NC or miR-204-5p inhibitor. b CCK-8 assays were performed to reveal the growth ability of SGC-7901 and AGS cells co-transfected with si-NC or si-DLX6-AS1 #3 and miR-NC or miR-204-5p inhibitor. c Transwell assays were performed to assess the invasion or migration function of SGC-7901 and AGS cells co-transfected with si-NC or si-DLX6-AS1 #3 and miR-NC or miR-204-5p inhibitor. d, e The mRNA and protein levels of EMT-related genes (E-cadherin, N-cadherin, SLUG and MMP9) in SGC-7901 and AGS cell lines co-transfected with si-NC or si-DLX6-AS1 #3 and miR-NC or miR-204-5p inhibitor were determined by qRT-PCR and western blotting. f F-actin staining assays were used to detect changes in the actin cytoskeleton in SGC-7901 and AGS cells co-transfected with si-NC or si-DLX6-AS1 #3 and miR-NC or miR-204-5p inhibitor. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01
Previous studies have indicated that miR-204-5p can regulate EMT in GC [25, 26]. Furthermore, we observed the morphological changes of GC cells and found that they developed a long fusiform and mesenchymal phenotype under miR-204-5p inhibitor treatment (Supplementary Fig. 5b). Therefore, we performed qRT-PCR and western blotting to analyze the EMT-related markers in co-transfected cell lines. As shown in Fig. 4d, e, miR-204-5p inhibitor promoted MMP9 and SLUG expression at transcriptional and translational levels (P < 0.05). Not only that, the tumor suppressor effect of si-DLX6-AS1 #3 was partially weakened by miR-204-5p inhibitor, including downregulation of E-cadherin and activation of SLUG and MMP9 at transcriptional and translational levels (P < 0.01). The results of phalloidin staining further indicated that suppression of miR-204-5p facilitated remodeling of the actin cytoskeleton and partially reversed the effect of si-DLX6-AS1 #3 by promoting F-actin expression and distribution (Fig. 4f).
OCT1 is a target gene of the DLX6-AS1/ miR-204-5p axis
To further clarify the oncogenic mechanism of the DLX6-AS1/miR-204-5p axis, we identified mRNAs that were positively co-expressed with DLX6-AS1 based on expression data from TCGA. As a result, we found a significant positive correlation between OCT1 and DLX6-AS1 in 375 GC samples (P = 3.22E−10, r = 0.659, Fig. 5a). According to the prediction of miRTarBase, the 3′-UTR region of OCT1 contains potential binding sites for miR-204-5p (Fig. 5b). We performed a luciferase reporter assay by co-transfecting with OCT1-WT or OCT1-MUT constructed plasmids and miR-204-5p mimic or miR-NC into the HEK293T, SGC-7901 and AGS cell lines. As shown in Fig. 5c, luciferase activity was significantly reduced in the OCT1-WT together with miR-204-5p mimics reaction system (P < 0.05). The results of qRT-PCR and western blotting showed that the expression of OCT1 could be promoted by miR-204-5p inhibitor (P < 0.01), but the effects could be weakened by co-transfection with si-DLX6-AS1 and miR-204-5p inhibitor (P < 0.01, Fig. 5d, e). As shown in Fig. 5f, there was a negative relationship between miR-204-5p and OCT1 expression from 56 GC tissues (P = 1.87E−4, r = − 0.472). Collectively, our results indicated that DLX6-AS1 positively regulates OCT1 expression by binding miR-204-5p in GC.
OCT1 is a target gene of the DLX6-AS1/ miR-204-5p axis. a Spearman’s correlation analysis of DLX6-AS1 expression and OCT1 expression in 375 GC patients from TCGA. b Binding sites between miR-204-5p and OCT1 were predicted using the bioinformatics database. c OCT1-WT vectors or OCT1-MUT vectors and miR-204-5p mimics or miR-NC were transfected into HEK293T, SGC-7901 and AGS cells, and the luciferase activity was measured. d The mRNA level of OCT1 in SGC-7901 and AGS cell lines co-transfected with si-NC or si-DLX6-AS1 #3 and miR-NC or miR-204-5p inhibitor were determined by qRT-PCR. e Protein levels of OCT1 in SGC-7901 and AGS cell lines treated with si-NC or si-DLX6-AS1 #3 and miR-NC or miR-204-5p inhibitor were determined by western blotting. f The correlation between miR-204-5p expression and OCT1 expression was measured in 56 GC patients by Spearman’s correlation analysis. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01
OCT1 is upregulated in GC and facilitates GC progression and the EMT in vitro
To investigate the function and mechanism of OCT1, we analyzed the expression of OCT1 in GC cell lines and tissues. According to the results of qRT-PCR and western blot assays, OCT1 was upregulated in GC as shown in Fig. 6a–c. Then, we transfected SGC-7901 and AGS cell lines with si-OCT1, and the transfection efficiency was confirmed by qRT-PCR and western blot assays (P < 0.01, Fig. 6d, e). Next, we explored whether OCT1 mediated the function of miR-204-5p. OCT1 downregulation inhibited proliferation of GC cells, and this effect could be partly reversed by the miR-204-5p inhibitor (P < 0.01, Fig. 6f, Supplementary Fig. 6a). Moreover, similar results were observed in Transwell assays (P < 0.01, Fig. 6g). GC cells exhibited relatively epithelial-like morphological features in the si-OCT1 group, and these were partly reversed by miR-204-4p downregulation (Supplementary Fig. 6b). As shown in Fig. 6h, i, knockdown of OCT1 reduced MMP9 and SLUG expression at both the transcriptional and translational levels (P < 0.01). However, the effect of si-OCT1 was partially weakened by miR-204-5p inhibitor, including inhibition of E-cadherin and promotion of SLUG as well as MMP9 (P < 0.01). Furthermore, we found that F-actin distribution and enrichment was significantly decreased under si-OCT1 treatment and also partly recovered by miR-204-5p inhibitor (Fig. 6j). Thus, the DLX6-AS1/miR-204-5p/OCT1 axis played an important role in GC progression and EMT.
OCT1 is upregulated in GC and facilitates GC progression and EMT in vitro. a, b OCT1 expression in a normal gastric cell line (GES-1) and five GC cell lines (SGC-7901, MCG-803, AGS, MKN-45 and HGC-27) was determined by qRT-PCR and western blotting. c The qRT-PCR analysis of OCT1 expression in 56 GC tissues and corresponding adjacent noncancerous tissues. Data are shown as − ΔΔCT values. d, e OCT1 expression in SGC-7901 and AGS cells transfected by si-NC and si-OCT1 was determined by qRT-PCR and western blotting. f CCK-8 assays were performed to reveal the growth ability of SGC-7901 and AGS cells transfected with si-NC, si-OCT1 #1 and miR-204-5p inhibitor. g Transwell assays were performed to assess the invasion or migration function of SGC-7901 and AGS cells transfected with si-NC, si-OCT1 #1 and miR-204-5p inhibitor. h, i mRNA and protein levels of EMT-related genes (E-cadherin, N-cadherin, SLUG and MMP9) in SGC-7901 and AGS cell lines transfected with si-NC, si-OCT1 #1 and miR-204-5p inhibitor were determined by qRT-PCR and western blotting. j F-actin staining assays were performed to detect changes in the actin cytoskeleton in SGC-7901 and AGS cells transfected with si-NC, si-OCT1 #1 and miR-204-5p inhibitor. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01
DLX6-AS1 is activated by the transcription factor OCT1 in GC
To further explore the regulatory mechanism of DLX6-AS1 in GC, we matched the promoter sequences of DLX6-AS1 with potential transcription factors using JASPAR. Interestingly, we found that the transcription factor OCT1 contained binding sites that could interact with the DLX6-AS1 promoter, and we chose the top five of them for further investigation (Fig. 7a, b). As shown in Fig. 7c, d, the P2 region (from − 750 to − 1350 bp) of the DLX6-AS1 promoter showed strong affinity to OCT1 by ChIP assays (P < 0.01). Moreover, we performed a luciferase assay to further investigate the specific binding sites between OCT1 and the DLX6-AS1 promoter. As shown in Fig. 7e, luciferase activity was elevated in WT and MUT2 of the P2 region group which was co-transfected with the OCT1 overexpression vector (P < 0.01). The results indicated that the sequence between − 928 and − 939 bp in the promoter is the validated binding site of OCT1. Furthermore, overexpression of OCT1 upregulated DLX6-AS1 in GC cell lines as shown in Fig. 7f (P < 0.01). According to qRT-PCR assays in 56 GC tissues, there was a positive correlation between OCT1 and DLX6-AS1 (P = 2.31E−8, r = 0.671, Fig. 7g). Hence, the results from our study revealed that DLX6-AS1/miR-204-5p/OCT1 formed a positive feedback loop in GC (Fig. 7h).
DLX6-AS1 is activated by transcription factor OCT1 in GC. a The binding motif of OCT1 from JASPAR tool. b, c The top five binding sites in the DLX6-AS1 promoter and their corresponding three counterparts in the promoter region are shown. d ChIP assays were performed to reveal the affinity of OCT1 to the P2 of the DLX6-AS1 promoter in SGC-7901 and AGS cells. e Luciferase reporter assays were used to locate the binding sequences of OCT1 to the DLX6-AS1 promoter. f Expression of DLX6-AS1 was detected in SGC-7901 and AGS cells transfected with OCT1 overexpression vector. g The correlation between DLX6-AS1 expression and OCT1 expression was measured in 56 GC patients by Spearman’s correlation analysis. h The mechanism of biological function of DLX6-AS1: OCT1-induced DLX6-AS1 promotes proliferation, metastasis, invasion and EMT processes of GC by inhibiting miR-204-5p and enhancing OCT1. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01
Discussion
Accumulating evidence has revealed that lncRNAs play a prominent role in the carcinogenesis and progression of GC. However, the regulatory mechanism and function of ectopic lncRNAs in cancer is still controversial. In our current study, we analyzed RNA-seq data from TCGA database and identified an upregulated and prognosis-related lncRNA, DLX6-AS1 which has not been explored in GC. According to qRT-PCR analysis, we confirmed that DLX6-AS1 was highly expressed in GC tissues and cell lines, and upregulation of DLX6-AS1 was associated with invasion and distant metastasis in GC. By knockdown of DLX6-AS1 in two different cell lines, we were able to restrain the proliferation, migration, and invasion of GC cells in vitro. Moreover, we found that DLX6-AS1 facilitated EMT accompanying actin cytoskeletal remodeling. It is worth noting that a recent study of DLX6-AS1 in pancreatic cancer has obtained similar results [18]. The EMT has been confirmed as a key mechanism that promotes invasion and immigration by inhibiting adhesion and reinforcing cell motility [27]. Nevertheless, the regulatory mechanisms of EMT still need to be understood, especially the effects involving lncRNAs and miRNAs. These findings demonstrated that DLX6-AS1 played an important role in GC and prompted us to conduct further mechanistic research.
In recent years, studies have demonstrated that lncRNAs function as ceRNAs to regulate expression of pro-cancer and anti-cancer genes in GC by sponging miRNAs. For instance, lncRNA KRTAP5-AS1 and lncRNA TUBB2A combined with miR-3620-3p and miR-596 constitute a regulatory network to further upregulate expression of Claudin-4 [28]. The lncRNA MIAT is upregulated in GC and increases DDX5 expression by targeting miR-141 [29]. In the present research, we performed bioinformatics analysis and experimental assays to identify and confirm that DLX6-AS1 could undergo crosstalk with miR-204-5p from four candidate miRNAs. The negative correlation between DLX6-AS1 and miR-204-5p was also analyzed and demonstrated in GC. Previous studies have reported that miR-204-5p is downregulated in GC and suppresses progression of tumors by multiple mechanisms. For instance, miR-204-5p targets SNAIL1 and regulates the EMT phenotype of GC [25]. Overexpression of miR-204-5p inhibits 5-FU resistance in GC cell lines by targeting TGFBR2 [26]. Consequently, it was imperative to identify the regulatory mechanism of miR-204-5p in GC. In this study, we revealed that miR-204-5p was expressed at low levels in GC cell lines and tissues, which was consistent with previous research. Not only that, but miR-204-5p also partially mediated the tumor-promoting function of DLX6-AS1 in GC progression and EMT.
Based on analysis of TCGA database, there was a positive correlation between expression of DLX6-AS1 and OCT1. Moreover, the 3′-UTR region of OCT1 contained potential binding sites for miR-204-5p. Then, we performed a series of experiments and verified the hypothesis that OCT1 is a target mRNA of the DLX6-AS1/miR-204-5p axis in GC. To the best of our knowledge, there has been no research into OCT1 regulation by ceRNAs in GC. OCT1 has been verified as a widely-expressed transcription factor in mammals and has been found to play an oncogenic role in GC and precancerous lesions [30]. In addition, previous studies reported that OCT1 could regulate EMT of diverse malignancies by targeting different signaling pathways [31,32,33]. Therefore, we performed rescue assays and confirmed that the DLX6-AS1/miR-204-5p/OCT1 axis was involved in the proliferation, migration, invasion and EMT of GC. Actin remodeling has been verified as the pivotal issue of the EMT. Previous studies mainly indicated that actin remodeling is the end result of EMT [34, 35], but recent research further reported that actin remodeling drives EMT by targeting HIF and Notch signaling [34]. Furthermore, an increasing number of lncRNAs and miRNAs have been found to participate in actin remodeling. For instance, Yu et al. [36] demonstrated that CXCL12 promoted progression of colorectal cancer through actin cytoskeletal remodeling by the lncRNA XIST/miR-133a-3p/RhoA axis. In the present research, we observed actin cytoskeleton remodeling through F-actin staining and first found that cytoskeletal organization was also regulated by the DLX6-AS1/miR-204-5p/OCT1 axis in GC. However, thorough research is needed to further reveal the regulatory mechanism of DLX6-AS1 between actin remodeling and EMT.
Although multiple studies have demonstrated that lncRNAs can be regulated by transcription factors in the cell nucleus [37,38,39], the regulatory mechanism of OCT1 in lncRNA regulation was still unknown. According to prediction by the JASPAR tool, there were potential binding sites between OCT1 and the DLX6-AS1 promoter and we hypothesized that transcription of DLX6-AS1 could be promoted by OCT1. Based on ChIP and luciferase reporter assays, we verified the hypothesis and located the binding sequences in the DLX6-AS1 promoter region. Furthermore, DLX6-AS1 could be significantly activated in vitro by transfecting with an OCT1 overexpression vector. These findings revealed a positive feedback loop (OCT1/DLX6-AS1/miR-204-5p/OCT1) which facilitated the progression and the EMT of GC.
In summary, our findings are the first to demonstrate that OCT1-induced DLX6-AS1 promoted GC proliferation, migration, invasion and EMT by sponging miR-204-5p and further upregulating OCT1. This present study revealed that DLX6-AS1 acted as a ceRNA in GC and provided a new conceivable prognostic biomarker and potential therapeutic target for GC patients.
Change history
09 November 2021
A Correction to this paper has been published: https://doi.org/10.1007/s10120-021-01263-9
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816.
Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7.
Ding F, Tang H, Nie D, Xia L. Long non-coding RNA Fer-1-like family member 4 is overexpressed in human glioblastoma and regulates the tumorigenicity of glioma cells. Oncol Lett. 2017;14:2379–84.
Wang X, Dong K, Jin Q, Ma Y, Yin S, Wang S. Upregulation of lncRNA FER1L4 suppresses the proliferation and migration of the hepatocellular carcinoma via regulating PI3K/AKT signal pathway. J Cell Biochem. 2019;120:6781–8.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8.
Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52.
Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C, et al. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16:123.
Dong S, Wang R, Wang H, Ding Q, Zhou X, Wang J, et al. HOXD-AS1 promotes the epithelial to mesenchymal transition of ovarian cancer cells by regulating miR-186-5p and PIK3R3. J Exp Clin Cancer Res. 2019;38:110.
Li J, Wang J, Chen Y, Li S, Jin M, Wang H, et al. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res. 2016;6:1099–107.
Liang C, Yang Y, Guan J, Lv T, Qu S, Fu Q, et al. LncRNA UCA1 sponges miR-204-5p to promote migration, invasion and epithelial-mesenchymal transition of glioma cells via upregulation of ZEB1. Pathol Res Pract. 2018;214:1474–81.
Li J, Li P, Zhao W, Yang R, Chen S, Bai Y, et al. Expression of long non-coding RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int. 2015;15:48.
Zhang L, He X, Jin T, Gang L, Jin Z. Long non-coding RNA DLX6-AS1 aggravates hepatocellular carcinoma carcinogenesis by modulating miR-203a/MMP-2 pathway. Biomed Pharmacother. 2017;96:884–91.
Li D, Tang X, Li M, Zheng Y. Long noncoding RNA DLX6-AS1 promotes liver cancer by increasing the expression of WEE1 via targeting miR-424-5p. J Cell Biochem. 2019;120:12290–9.
Zeng X, Hu Z, Ke X, Tang H, Wu B, Wei X, et al. Long noncoding RNA DLX6-AS1 promotes renal cell carcinoma progression via miR-26a/PTEN axis. Cell Cycle. 2017;16:2212–9.
Zhang RM, Tang T, Yu HM, Yao XD. LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem Biophys Res Commun. 2018;507:260–6.
Li X, Zhang H, Wu X. Long noncoding RNA DLX6-AS1 accelerates the glioma carcinogenesis by competing endogenous sponging miR-197-5p to relieve E2F1. Gene. 2018;686:1–7.
An Y, Chen XM, Yang Y, Mo F, Jiang Y, Sun DL, et al. LncRNA DLX6-AS1 promoted cancer cell proliferation and invasion by attenuating the endogenous function of miR-181b in pancreatic cancer. Cancer Cell Int. 2018;18:143.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8.
Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–3.
Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–8.
Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302.
Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D1284.
Du Y, Jiang B, Song S, Pei G, Ni X, Wu J, et al. Metadherin regulates actin cytoskeletal remodeling and enhances human gastric cancer metastasis via epithelial-mesenchymal transition. Int J Oncol. 2017;51:63–74.
Liu Z, Long J, Du R, Ge C, Guo K, Xu Y. miR-204 regulates the EMT by targeting snai1 to suppress the invasion and migration of gastric cancer. Tumor Biol. 2016;37:8327–35.
Li LQ, Pan D, Chen Q, Zhang SW, Xie DY, Zheng XL, et al. Sensitization of gastric cancer cells to 5-FU by microRNA-204 through targeting the TGFBR2-mediated epithelial to mesenchymal transition. Cell Physiol Biochem. 2018;47:1533–45.
Peng Z, Wang CX, Fang EH, Wang GB, Tong Q. Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J Gastroenterol. 2014;20:5403–10.
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX, Wu ZH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289.
Sha M, Lin M, Wang J, Ye J, Xu J, Xu N, et al. Long non-coding RNA MIAT promotes gastric cancer growth and metastasis through regulation of miR-141/DDX5 pathway. J Exp Clin Cancer Res. 2018;37:58.
Qian J, Kong X, Deng N, Tan P, Chen H, Wang J, et al. OCT1 is a determinant of synbindin-related ERK signalling with independent prognostic significance in gastric cancer. Gut. 2015;64:37–48.
Hwang-Verslues WW, Chang PH, Jeng YM, Kuo WH, Chiang PH, Chang YC, et al. Loss of corepressor PER2 under hypoxia up-regulates OCT1-mediated EMT gene expression and enhances tumor malignancy. Proc Natl Acad Sci USA. 2013;110:12331–6.
Xu SH, Huang JZ, Xu ML, Yu G, Yin XF, Chen D, et al. ACK1 promotes gastric cancer epithelial-mesenchymal transition and metastasis through AKT-POU2F1-ECD signalling. J Pathol. 2015;236:175–85.
Zhong Y, Huang H, Chen M, Huang J, Wu Q, Yan GR, et al. POU2F1 over-expression correlates with poor prognoses and promotes cell growth and epithelial-to-mesenchymal transition in hepatocellular carcinoma. Oncotarget. 2017;8:44082–95.
Peng JM, Bera R, Chiou CY, Yu MC, Chen TC, Chen CW, et al. Actin cytoskeleton remodeling drives epithelial-mesenchymal transition for hepatoma invasion and metastasis in mice. Hepatology. 2018;67:2226–43.
Dai J, Qin L, Chen Y, Wang H, Lin G, Li X, et al. Matrix stiffness regulates epithelial-mesenchymal transition via cytoskeletal remodeling and MRTF-A translocation in osteosarcoma cells. J Mech Behav Biomed Mater. 2019;90:226–38.
Yu X, Wang D, Wang X, Sun S, Zhang Y, Wang S, et al. CXCL12/CXCR36 promotes inflammation-driven colorectal cancer progression through activation of RhoA signaling by sponging miR-133a-3p. J Exp Clin Cancer Res. 2019;38:32.
Fan W, Peng Y, Liang Z, Yang Y, Zhang J. A negative feedback loop of H19/miR-675/EGR1 is involved in diabetic nephropathy by downregulating the expression of the vitamin D receptor. J Cell Physiol. 2019;234:17505–13.
Bian Y, Gao G, Zhang Q, Qian H, Yu L, Yao N, et al. KCNQ1OT1/miR-217/ZEB1 feedback loop facilitates cell migration and epithelial-mesenchymal transition in colorectal cancer. Cancer Biol Ther. 2019;20:886–96.
Ma F, Liu X, Zhou S, Li W, Liu C, Chadwick M, et al. Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 2019;450:63–75.
Funding
The study was funded by the National Natural Science Foundation of China (Grant number 30572162); the Natural Science Foundation of Liaoning Province (Grant number 201602817); Chun-Dong Zhang was supported in part by the Japan China Sasakawa Medical Fellowship (Grant number 2017816).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Yu Liang declares that he has no conflict of interest. Chun-Dong Zhang declares that he has no conflict of interest. Cheng Zhang declares that he has no conflict of interest. Dong-Qiu Dai declares that he has no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liang, Y., Zhang, CD., Zhang, C. et al. DLX6-AS1/miR-204-5p/OCT1 positive feedback loop promotes tumor progression and epithelial–mesenchymal transition in gastric cancer. Gastric Cancer 23, 212–227 (2020). https://doi.org/10.1007/s10120-019-01002-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-019-01002-1