Abstract
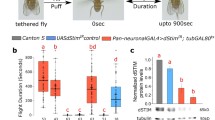
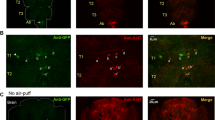
Mutants in the Drosophila InsP3R gene (itpr) are flight defective. Expression of the wild-type InsP3R in aminergic interneurons rescues flight. However, molecular and cellular changes in the central nervous system of InsP3R mutants that lead to flightless behavior remain unknown. To understand the molecular basis of flight phenotypes in Drosophila InsP3R mutants a microarray screen was done with RNA isolated from adult heads and thoraces. We found down-regulation of several genes that affect the excitability of neurons and muscles. Among these the role of glutamine synthetase 2 was investigated further. This enzyme reduces glutamate levels at the synapse. Our results show that Gs2 (glutamine synthetase 2) transcripts and glutamate levels correspond precisely but inversely, with InsP3R phenotypes, which can be rescued by a mutant allele for Gs2, namely, Gs2 m13. From measuring axonal branches and synapse number at a glutamatergic synapse—the adult flight neuromuscular junction—of InsP3R mutants, we conclude that glutamate homeostasis is altered through a cell non-autonomous mechanism, and is likely to be an important basis for the observed flight defects.








Similar content being viewed by others
References
Banerjee S, Lee J, Venkatesh K, Wu CF, Hasan G (2004) Loss of flight and associated neuronal rhythmicity in inositol 1,4,5-trisphosphate receptor mutants of Drosophila. J Neurosci 24:7869–7878
Banerjee S, Joshi R, Venkiteswaran G, Agrawal N, Srikanth S, Alam F, Hasan G (2006) Compensation of inositol 1,4,5-trisphosphate receptor function by altering sarco-endoplasmic reticulum calcium ATPase activity in the Drosophila flight circuit. J Neurosci 26:8278–8288
Behra M, Cousin X, Bertrand C, Vonesch JL, Biellmann D, Chatonnet A, Strahle U (2002) Acetylcholinesterase is required for neuronal and muscular development in the zebrafish embryo. Nat Neurosci 5:111–118
Benzer S (1973) Genetic dissection of behavior. Sci Am 229:24–37
Berridge MJ, Bootman MD, Roderick HL (2003) Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol 4:517–529
Borodinsky LN, Root CM, Cronin JA, Sann SB, Gu X, Spitzer NC (2004) Activity-dependent homeostatic specification of transmitter expression in embryonic neurons. Nature 429:523–530
Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415
Brandon EP, Lin W, D’Amour KA, Pizzo DP, Dominguez B, Sugiura Y, Thode S, Ko CP, Thal LJ, Gage FH, Lee KF (2003) Aberrant patterning of neuromuscular synapses in choline acetyltransferase-deficient mice. J Neurosci 23:539–549
Budnik V, Wu CF, White K (1989) Altered branching of serotonin-containing neurons in Drosophila mutants unable to synthesize serotonin and dopamine. J Neurosci 9:2866–2877
Budnik V, Zhong Y, Wu CF (1990) Morphological plasticity of motor axons in Drosophila mutants with altered excitability. J Neurosci 10:3754–3768
Chiang PW, Kurnit DM (2003) Study of dosage compensation in Drosophila. Genetics 165:1167–1181
Ciccolini F, Collins TJ, Sudhoelter J, Lipp P, Berridge MJ, Bootman MD (2003) Local and global spontaneous calcium events regulate neurite outgrowth and onset of GABAergic phenotype during neural precursor differentiation. J Neurosci 23:103–111
Consoulas C, Restifo LL, Levine RB (2002) Dendritic remodeling and growth of motoneurons during metamorphosis of Drosophila melanogaster. J Neurosci 22:4906–4917
Cooper RL, Neckameyer WS (1999) Dopaminergic modulation of motor neuron activity and neuromuscular function in Drosophila melanogaster. Comp Biochem Physiol B Biochem Mol Biol 122:199–210
Dasari S, Cooper RL (2004) Modulation of sensory-CNS-motor circuits by serotonin, octopamine, and dopamine in semi-intact Drosophila larva. Neurosci Res 48:221–227
Engel JE, Wu CF (1996) Altered habituation of an identified escape circuit in Drosophila memory mutants. J Neurosci 16:3486–3499
Featherstone DE, Rushton E, Broadie K (2002) Developmental regulation of glutamate receptor field size by nonvesicular glutamate release. Nat Neurosci 5:141–146
Fernandes JJ, Keshishian H (1999) Development of the adult neuromuscular system. Int Rev Neurobiol 43:221–239
Ganetzky B, Wu CF (1983) Neurogenetic analysis of potassium currents in Drosophila: synergistic effects on neuromuscular transmission in double mutants. J Neurogenet 1:17–28
Harden N, Lee J, Loh HY, Ong YM, Tan I, Leung T, Manser E, Lim L (1996) A Drosophila homolog of the Rac- and Cdc42-activated serine/threonine kinase PAK is a potential focal adhesion and focal complex protein that colocalizes with dynamic actin structures. Mol Cell Biol 16:1896–1908
Hebbar S, Fernandes JJ (2004) Pruning of motor neuron branches establishes the DLM innervation pattern in Drosophila. J Neurobiol 60:499–516
Inoue T, Kato K, Kohda K, Mikoshiba K (1998) Type 1 inositol 1,4,5-trisphosphate receptor is required for induction of long-term depression in cerebellar Purkinje neurons. J Neurosci 18:5366–5373
Jhaveri D, Sen A, Reddy GV, Rodrigues V (2000) Sense organ identity in the Drosophila antenna is specified by the expression of the proneural gene atonal. Mech Dev 99:101–111
Joshi R, Venkatesh K, Srinivas R, Nair S, Hasan G (2004) Genetic dissection of itpr gene function reveals a vital requirement in aminergic cells of Drosophila larvae. Genetics 166:225–236
LaFerla FM (2002) Calcium dyshomeostasis and intracellular signalling in Alzheimer’s disease. Nat Rev Neurosci 3:862–872
Lee J, Wu CF (2002) Electroconvulsive seizure behavior in Drosophila: analysis of the physiological repertoire underlying a stereotyped action pattern in bang-sensitive mutants. J Neurosci 22:11065–11079
Li H, Chaney S, Roberts IJ, Forte M, Hirsh J (2000) Ectopic G-protein expression in dopamine and serotonin neurons blocks cocaine sensitization in Drosophila melanogaster. Curr Biol 10:211–214
Lin X, Antalffy B, Kang D, Orr HT, Zoghbi HY (2000) Polyglutamine expansion down-regulates specific neuronal genes before pathologic changes in SCA1. Nat Neurosci 3:157–163
Lorentzos P, Kaiser T, Kennerson ML, Nicholson GA (2003) A rapid and definitive test for Charcot-Marie-Tooth 1A and hereditary neuropathy with liability to pressure palsies using multiplexed real-time PCR. Genet Test 7:135–138
Saitoe M, Schwarz TL, Umbach JA, Gundersen CB, Kidokoro Y (2001) Absence of junctional glutamate receptor clusters in Drosophila mutants lacking spontaneous transmitter release. Science 293:514–517
Sanyal S, Consoulas C, Kuromi H, Basole A, Mukai L, Kidokoro Y, Krishnan KS, Ramaswami M (2005) Analysis of conditional paralytic mutants in Drosophila sarco-endoplasmic reticulum calcium ATPase reveals novel mechanisms for regulating membrane excitability. Genetics 169:737–750
Sone M, Suzuki E, Hoshino M, Hou D, Kuromi H, Fukata M, Kuroda S, Kaibuchi K, Nabeshima Y, Hama C (2000) Synaptic development is controlled in the periactive zones of Drosophila synapses. Development 127:4157–4168
Srikanth S, Wang Z, Tu H, Nair S, Mathew MK, Hasan G, Bezprozvanny I (2004) Functional properties of the Drosophila melanogaster inositol 1,4,5-trisphosphate receptor mutants. Biophys J 86:3634–3646
Tang TS, Tu H, Chan EY, Maximov A, Wang Z, Wellington CL, Hayden MR, Bezprozvanny I (2003) Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron 39:227–239
Tanouye MA, Wyman RJ (1980) Motor outputs of giant nerve fiber in Drosophila. J Neurophysiol 44:405–421
Venkatesh K, Siddhartha G, Joshi R, Patel S, Hasan G (2001) Interactions between the inositol 1,4,5-trisphosphate and cyclic AMP signaling pathways regulate larval molting in Drosophila. Genetics 158:309–318
Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E (2006) Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49:833–844
Acknowledgments
We thank V. Karthikeyan and S. Banerjee for discussions and help in analysis of the data. We also thank Dr. H. Krishnamurthy and the imaging facility at NCBS for their help and expertise. SN is supported by a senior research fellowship from the Department of Science and Technology. This work was supported by a core grant from the National Centre for Biological Sciences, TIFR, Bangalore.
Author information
Authors and Affiliations
Corresponding author
Supplementary materials and methods
Total RNA preparation, cDNA labeling and array hybridization Total RNA was extracted from approximately 200 fly heads and thoraces using standard procedures and then subjected to DNAse treatment. The RNA was purified using the RNeasy minikit (Qiagen). For quality assurance, the RNA was visualized on a TBE gel. Approximately 65-80 μg of total RNA from wild-type and mutant samples was separately reverse-transcribed using the MicroMax™ Direct cDNA Labeling Kit (Perkin Elmer Life Sciences, Boston, MA, USA) into Cy3 and Cy5 labeled cDNA targets respectively. Labeled control and mutant cDNAs were pooled and purified using Microcon YM-100 filter units (Millipore Corp., Bedford, MA, USA). Equal efficiency of labeling was verified in both reactions by running the labeled cDNA on a 0.8% agarose gel cast on a slide and scanned with the GeneTAC LS IV Scanner from Genomic Solutions. Microarray experiments were performed using the Drosophila 7K array V2.0 (Microarray services, University of Toronto, Canada) containing 16,896 spots (including controls and blank spots). The slide contained 7,378 independent genes from the Drosophila genome spotted in duplicates. Each experiment compared RNA samples from wild-type and mutant flies. For each genotype, experiments were repeated at least 3 times or more using independently isolated RNA. After subjecting the slides to prehybridization at 42°C for 45 min and washing with isopropanol and MilliQ double distilled water, hybridization was carried out in the GeneTACTM Hybridization Station (Genomic Solutions) at 42°C for 16 hours. Hybridised slides were washed attachment to manuscript Click here to download attachment to manuscript: Supplementary.doc using medium stringency, high stringency and post-wash buffers for 20 sec each, dried and scanned using the GeneTACTM LS IV scanner (Genomic Solutions).
Microarray Image and Data Analysis
Signals generated from Cy3 and Cy5 channels on each microarray were background subtracted and normalized to the total signals of all spots, and analyzed using GeneTac Analyzer V3.3. (Genomic Solutions) and Excel (Microsoft). The following stringent criteria were used to select spots with reliable and optimal fluorescence intensity for further analysis. 1) In independent experiments, only spots with a signal-to-noise ratio greater than blank spots were taken for further analysis. 2) If the coefficient of variation of ratios of duplicate spots of a given gene was equal or less than 20%, it was taken for further analysis. 3) The ratio of Cy5 to Cy3 flourescence intensity for each gene was normalized using the median Cy5/Cy3 ratio of the rp49 spots. The normalized data were subjected to hierarchical Cluster analysis using Gene Cluster, version 2.11 and visually represented using Treeview (software developed by Michael Eisen, Stanford University). Data were represented as a fold change of fluorescence intensity of a gene from test sample versus reference sample. Anegative fold change ≥ 1.7 was taken to represent down-regulation of the gene. Average values and standard deviation for triplicate experiments were determined. Genes were considered to be differentially expressed only if they displayed the same trend of change in expression in each of the triplicate experiments. Gene annotation information was obtained from Flybase.
10158_2007_48_MOESM1_ESM.tif
Supplementary Fig. S1A. Down regulated gene transcription profiles from the heads and thoraces of two InsP3R mutants (itpr), ka1091/ug3 and wc703/ug3. List of genes that are down-regulated in ka1091/ug3 and wc703/ug3 head tissue. The color image on the left is a visual representation of the fold downregulation in both the genotypes. Results shown are average of data obtained from at least three independent experiments. Gene names are from Flybase (http://flybase.bio.indiana.edu/). (TIF 2.42 MB)
10158_2007_48_MOESM2_ESM.tif
Supplementary Fig. S1B. Down regulated gene transcription profiles from the heads and thoraces of two InsP3R mutants (itpr), ka1091/ug3 and wc703/ug3. List of genes that are down-regulated in ka1091/ug3 and wc703/ug3 thoracic tissue. (TIF 1.23 MB)
10158_2007_48_MOESM3_ESM.tif
Supplementary Fig. S2. Representative scatter plots of log (base 10) of Cy3 and Cy5 signal intensities from hybridization of probes obtained from independent RNA samples. Each point represents the normalized Cy3 and Cy5 intensity values of genes spotted. The Cy3 and Cy5 intensities were normalized based on the Cy5/Cy3 ratio of the control gene, rp49. Here, most of the data points lie on the diagonal line indicating that a majority of the genes are not regulated. Spots that lie farther away from the diagonal correspond to genes that are differentially expressed. In the slide hybridized to ka1091/ug3 head RNA, among the 7378 correctly spotted genes, 6278 genes showed no change in their expression level while approximately 1100 genes showed altered expression levels. After further stringent statistical analysis, based on the signal-noise ratio of the blank spots and positive controls and the coefficient of variation of the duplicate spots, this number was reduced to approximately 300 genes. Similarly, in the slide hybridized to RNA from ka1091/ug3 thorax, 6120 genes showed normal expression levels while approximately 1258 genes were differentially expressed. Among these, 418 genes were considered for further analysis. The data points are color coded such that spots that are induced > 1 fold are colored red and those induced < 1 are colored in green. The genes that appear up-regulated in this experiment were not reproduced in more than two experiments. (TIF 931 kB)
10158_2007_48_MOESM4_ESM.tif
Supplementary Fig. S3. Genomic DNA PCR of CS and Gs2 m13/+. Genomic DNA extracted from female virgins of CS and Gs2 m13/+ were analyzed by PCR in order to quantify the level of Gs2 genomic DNA. The positive control used was a region in the itpr gene. Gs2 DNA in Gs2 m13/+ was found to be approximately 1.9 fold reduced as compared with CS. itpr gene levels were not different in CS and Gs2 m13/+. (TIF 69.1 kB)
10158_2007_48_MOESM5_ESM.avi
Supplementary Movie 1. Three-dimensional projection of an arbor at a secondary contact point on a ka1091/ug3 mutant (Movie 1) and Canton-S (Movie 2) DLM. 22C10 staining is in green and Anti-DPAK staining is in red. The movies correspond to the images shown in Fig. 4 A-H (ka1091/ug3 image has been rotated by 180° in the figure). The movies show a series of 6 projections at a difference angle of 6° rotating along the y-axis at a speed of 3 frames per second. For the actual analysis, a series of 64 projections at a difference angle of 6° along the y-axis were used. (AVI 12.3 MB)
10158_2007_48_MOESM6_ESM.avi
Supplementary Movie 2. Three-dimensional projection of an arbor at a secondary contact point on a ka1091/ug3 mutant (Movie 1) and Canton-S (Movie 2) DLM. 22C10 staining is in green and Anti-DPAK staining is in red. The movies correspond to the images shown in Fig. 4 A-H (ka1091/ug3 image has been rotated by 180° in the figure). The movies show a series of 6 projections at a difference angle of 6° rotating along the y-axis at a speed of 3 frames per second. For the actual analysis, a series of 64 projections at a difference angle of 6° along the y-axis were used. (AVI 12.6 MB)
Rights and permissions
About this article
Cite this article
Nair, S., Agrawal, N. & Hasan, G. Homeostasis of glutamate neurotransmission is altered in Drosophila Inositol 1,4,5-trisphosphate receptor mutants. Invert Neurosci 7, 137–147 (2007). https://doi.org/10.1007/s10158-007-0048-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10158-007-0048-0