Abstract
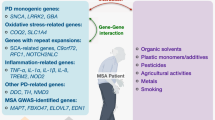
Classically defined phenotypically by a triad of cerebellar ataxia, parkinsonism, and autonomic dysfunction in conjunction with pyramidal signs, multiple system atrophy (MSA) is a rare and progressive neurodegenerative disease affecting an estimated 3–4 per every 100,000 individuals among adults 50–99 years of age. With a pathological hallmark of alpha-synuclein-immunoreactive glial cytoplasmic inclusions (GCIs; Papp–Lantos inclusions), MSA patients exhibit marked neurodegenerative changes in the striatonigral and/or olivopontocerebellar structures of the brain. As a member of the alpha-synucleinopathy family, which is defined by its well-demarcated alpha-synuclein-immunoreactive inclusions and aggregation, MSA’s clinical presentation exhibits several overlapping features with other members including Parkinson’s disease (PD) and dementia with Lewy bodies (DLB). Given the extensive fund of knowledge regarding the genetic etiology of PD revealed within the past several years, a genetic investigation of MSA is warranted. While a current genome-wide association study is underway for MSA to further clarify the role of associated genetic loci and single-nucleotide polymorphisms, several cases have presented solid preliminary evidence of a genetic etiology. Naturally, genes and variants manifesting known associations with PD (and other phenotypically similar neurodegenerative disorders), including SNCA and MAPT, have been comprehensively investigated in MSA patient cohorts. More recently variants in COQ2 have been linked to MSA in the Japanese population although this finding awaits replication. Nonetheless, significant positive associations with subsequent independent replication studies have been scarce. With very limited information regarding genetic mutations or alterations in gene dosage as a cause of MSA, the search for novel risk genes, which may be in the form of common variants or rare variants, is the logical nexus for MSA research. We believe that the application of next generation genetic methods to MSA will provide valuable insight into the underlying causes of this disease, and will be central to the identification of etiologic-based therapies.
Similar content being viewed by others
References
Ahmed Z, Asi YT, Sailer A et al (2012) The neuropathology, pathophysiology and genetics of multiple system atrophy. Neuropathol Appl Neurobiol 38:4–24. doi:10.1111/j.1365-2990.2011.01234.x
Stefanova N, Bücke P, Duerr S, Wenning GK (2009) Multiple system atrophy: an update. Lancet Neurol 8:1172–1178. doi:10.1016/S1474-4422(09)70288-1
Bower JH, Maraganore DM, McDonnell SK, Rocca WA (1997) Incidence of progressive supranuclear palsy and multiple system atrophy in Olmsted County, Minnesota, 1976 to 1990. Neurology 49:1284–1288
Wüllner U, Abele M, Schmitz-Huebsch T et al (2004) Probable multiple system atrophy in a German family. J Neurol Neurosurg Psychiatry 75:924–925
Osaki Y, Wenning GK, Daniel SE et al (2002) Do published criteria improve clinical diagnostic accuracy in multiple system atrophy? Neurology 59:1486–1491
Scholz SW, Houlden H, Schulte C et al (2009) SNCA variants are associated with increased risk for multiple system atrophy. Ann Neurol 65:610–614. doi:10.1002/ana.21685
Gilman S, Wenning GK, Low PA et al (2008) Second consensus statement on the diagnosis of multiple system atrophy. Neurology 71:670–676. doi:10.1212/01.wnl.0000324625.00404.15
Hara K, Momose Y, Tokiguchi S et al (2007) Multiplex families with multiple system atrophy. Arch Neurol 64:545–551. doi:10.1001/archneur.64.4.545
Kiely AP, Asi YT, Kara E et al (2013) α-Synucleinopathy associated with G51D SNCA mutation: a link between Parkinson’s disease and multiple system atrophy? Acta Neuropathol (Berl) 125:753–769. doi:10.1007/s00401-013-1096-7
Yoshida M (2011) Multiple system atrophy—synuclein and neuronal degeneration. Rinshō Shinkeigaku Clin Neurol 51:838–842
Ozawa T, Tada M, Kakita A et al (2010) The phenotype spectrum of Japanese multiple system atrophy. J Neurol Neurosurg Psychiatry 81:1253–1255. doi:10.1136/jnnp.2009.182576
Wenning GK, Wagner S, Daniel S, Quinn NP (1993) Multiple system atrophy: sporadic or familial? Lancet 342:681
Vanacore N (2005) Epidemiological evidence on multiple system atrophy. J Neural Transm 112:1605–1612. doi:10.1007/s00702-005-0380-7
Fogel BL, Clark MC, Geschwind DH (2014) The neurogenetics of atypical parkinsonian disorders. Semin Neurol 34:217–224. doi:10.1055/s-0034-1381738
Stamelou M, Quinn NP, Bhatia KP (2013) “Atypical” atypical parkinsonism: new genetic conditions presenting with features of progressive supranuclear palsy, corticobasal degeneration, or multiple system atrophy—a diagnostic guide. Mov Disord Off J Mov Disord Soc. doi:10.1002/mds.25509
Wenning GK, Geser F, Krismer F et al (2013) The natural history of multiple system atrophy: a prospective European cohort study. Lancet Neurol 12:264–274. doi:10.1016/S1474-4422(12)70327-7
Gilman S, Sima AA, Junck L et al (1996) Spinocerebellar ataxia type 1 with multiple system degeneration and glial cytoplasmic inclusions. Ann Neurol 39:241–255. doi:10.1002/ana.410390214
Nirenberg MJ, Libien J, Vonsattel J-P, Fahn S (2007) Multiple system atrophy in a patient with the spinocerebellar ataxia 3 gene mutation. Mov Disord Off J Mov Disord Soc 22:251–254. doi:10.1002/mds.21231
Huang Y, Hayes M, Harding AJ et al (2006) Anticipation of onset age in familial Parkinson’s disease without SCA gene mutations. Parkinsonism Relat Disord 12:309–313. doi:10.1016/j.parkreldis.2006.01.002
Schöls L, Bauer P, Schmidt T et al (2004) Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol 3:291–304. doi:10.1016/S1474-4422(04)00737-9
Khan NL, Giunti P, Sweeney MG et al (2005) Parkinsonism and nigrostriatal dysfunction are associated with spinocerebellar ataxia type 6 (SCA6). Mov Disord Off J Mov Disord Soc 20:1115–1119. doi:10.1002/mds.20564
Kim J-Y, Kim SY, Kim J-M et al (2009) Spinocerebellar ataxia type 17 mutation as a causative and susceptibility gene in parkinsonism. Neurology 72:1385–1389. doi:10.1212/WNL.0b013e3181a18876
Abele M, Bürk K, Schöls L et al (2002) The aetiology of sporadic adult-onset ataxia. Brain J Neurol 125:961–968
Lin I-S, Wu R-M, Lee-Chen G-J et al (2007) The SCA17 phenotype can include features of MSA-C, PSP and cognitive impairment. Parkinsonism Relat Disord 13:246–249. doi:10.1016/j.parkreldis.2006.04.009
Kim H-J, Jeon BS, Shin J et al (2014) Should genetic testing for SCAs be included in the diagnostic workup for MSA? Neurology 83:1733–1738. doi:10.1212/WNL.0000000000000965
Stemberger S, Scholz SW, Singleton AB, Wenning GK (2011) Genetic players in multiple system atrophy: unfolding the nature of the beast. Neurobiol Aging 32(1924):e5–e14. doi:10.1016/j.neurobiolaging.2011.04.001
Stemberger S, Wenning GK (2011) Modelling progressive autonomic failure in MSA: where are we now? J Neural Transm 118:841–847. doi:10.1007/s00702-010-0576-3
Fernagut P-O, Tison F (2012) Animal models of multiple system atrophy. Neuroscience 211:77–82. doi:10.1016/j.neuroscience.2011.09.044
Flabeau O, Meissner WG, Tison F (2010) Multiple system atrophy: current and future approaches to management. Ther Adv Neurol Disord 3:249–263. doi:10.1177/1756285610375328
Wenning GK, Ben-Shlomo Y, Hughes A et al (2000) What clinical features are most useful to distinguish definite multiple system atrophy from Parkinson’s disease? J Neurol Neurosurg Psychiatry 68:434–440
Ozawa T, Paviour D, Quinn NP et al (2004) The spectrum of pathological involvement of the striatonigral and olivopontocerebellar systems in multiple system atrophy: clinicopathological correlations. Brain J Neurol 127:2657–2671. doi:10.1093/brain/awh303
Kisos H, Pukaß K, Ben-Hur T et al (2012) Increased neuronal α-synuclein pathology associates with its accumulation in oligodendrocytes in mice modeling α-synucleinopathies. PLoS One 7:e46817. doi:10.1371/journal.pone.0046817
Rockenstein E, Ubhi K, Inglis C et al (2012) Neuronal to oligodendroglial α-synuclein redistribution in a double transgenic model of multiple system atrophy. Neuroreport 23:259–264. doi:10.1097/WNR.0b013e3283509842
Yoshida M (2007) Multiple system atrophy: alpha-synuclein and neuronal degeneration. Neuropathol Off J Jpn Soc Neuropathol 27:484–493
Kato S, Shinozawa T, Takikawa M et al (2000) Midkine, a new neurotrophic factor, is present in glial cytoplasmic inclusions of multiple system atrophy brains. Acta Neuropathol (Berl) 100:481–489
Muramatsu T (1992) Retinoic acid regulates the expression of a new heparin binding growth differentiation factor. J Nutr Sci Vitaminol (Tokyo) Spec No:485–487
Nurcombe V, Fraser N, Herlaar E, Heath JK (1992) MK: a pluripotential embryonic stem-cell-derived neuroregulatory factor. Dev Camb Engl 116:1175–1183
Satoh J, Muramatsu H, Moretto G et al (1993) Midkine that promotes survival of fetal human neurons is produced by fetal human astrocytes in culture. Brain Res Dev Brain Res 75:201–205
Ishizawa K, Komori T, Sasaki S et al (2004) Microglial activation parallels system degeneration in multiple system atrophy. J Neuropathol Exp Neurol 63:43–52
Lehotzky A, Lau P, Tokési N et al (2010) Tubulin polymerization-promoting protein (TPPP/p25) is critical for oligodendrocyte differentiation. Glia 58:157–168. doi:10.1002/glia.20909
Hasegawa T, Baba T, Kobayashi M et al (2010) Role of TPPP/p25 on α-synuclein-mediated oligodendroglial degeneration and the protective effect of SIRT2 inhibition in a cellular model of multiple system atrophy. Neurochem Int 57:857–866. doi:10.1016/j.neuint.2010.09.002
Song YJC, Lundvig DMS, Huang Y et al (2007) p25alpha relocalizes in oligodendroglia from myelin to cytoplasmic inclusions in multiple system atrophy. Am J Pathol 171:1291–1303. doi:10.2353/ajpath.2007.070201
Sugeno N, Takeda A, Hasegawa T et al (2008) Serine 129 phosphorylation of alpha-synuclein induces unfolded protein response-mediated cell death. J Biol Chem 283:23179–23188. doi:10.1074/jbc.M802223200
Riedel M, Goldbaum O, Wille M, Richter-Landsberg C (2011) Membrane lipid modification by docosahexaenoic acid (DHA) promotes the formation of α-synuclein inclusion bodies immunopositive for SUMO-1 in oligodendroglial cells after oxidative stress. J Mol Neurosci MN 43:290–302. doi:10.1007/s12031-010-9439-5
Outeiro TF, Kontopoulos E, Altmann SM et al (2007) Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 317:516–519. doi:10.1126/science.1143780
Ozawa T, Revesz T, Paviour D et al (2012) Difference in MSA phenotype distribution between populations: genetics or environment? J Park Dis 2:7–18. doi:10.3233/JPD-2012-11056
Nee LE, Gomez MR, Dambrosia J et al (1991) Environmental-occupational risk factors and familial associations in multiple system atrophy: a preliminary investigation. Clin Auton Res Off J Clin Auton Res Soc 1:9–13
Vanacore N, Bonifati V, Fabbrini G et al (2005) Case-control study of multiple system atrophy. Mov Disord Off J Mov Disord Soc 20:158–163. doi:10.1002/mds.20303
Davidson WS, Jonas A, Clayton DF, George JM (1998) Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem 273:9443–9449
Lee PH, Lim TS, Shin H-W et al (2009) Serum cholesterol levels and the risk of multiple system atrophy: a case-control study. Mov Disord Off J Mov Disord Soc 24:752–758. doi:10.1002/mds.22459
Armstrong RA, Cairns NJ, Lantos PL (2006) Multiple system atrophy (MSA): topographic distribution of the alpha-synuclein-associated pathological changes. Parkinsonism Relat Disord 12:356–362. doi:10.1016/j.parkreldis.2006.02.005
Vidal J-S, Vidailhet M, Derkinderen P et al (2010) Familial aggregation in atypical Parkinson’s disease: a case control study in multiple system atrophy and progressive supranuclear palsy. J Neurol 257:1388–1393. doi:10.1007/s00415-010-5638-9
Multiple-System Atrophy Research Collaboration (2013) Mutations in COQ2 in familial and sporadic multiple-system atrophy. N Engl J Med 369:233–244. doi:10.1056/NEJMoa1212115
Jeon BS, Farrer MJ, Bortnick SF, Korean Canadian Alliance on Parkinson’s Disease and Related Disorders (2014) Mutant COQ2 in multiple-system atrophy. N Engl J Med 371:80. doi:10.1056/NEJMc1311763#SA1
Sharma M, Wenning G, Krüger R, European Multiple-System Atrophy Study Group (EMSA-SG) (2014) Mutant COQ2 in multiple-system atrophy. N Engl J Med 371:80–81. doi:10.1056/NEJMc1311763#SA2
Schottlaender LV, Houlden H, Multiple-System Atrophy (MSA) Brain Bank Collaboration (2014) Mutant COQ2 in multiple-system atrophy. N Engl J Med 371:81. doi:10.1056/NEJMc1311763#SA3
Bleasel JM, Wong JH, Halliday GM, Kim WS (2014) Lipid dysfunction and pathogenesis of multiple system atrophy. Acta Neuropathol Commun 2:15. doi:10.1186/2051-5960-2-15
Soma H, Yabe I, Takei A et al (2008) Associations between multiple system atrophy and polymorphisms of SLC1A4, SQSTM1, and EIF4EBP1 genes. Mov Disord Off J Mov Disord Soc 23:1161–1167. doi:10.1002/mds.22046
Wyss-Coray T, Mucke L (2002) Inflammation in neurodegenerative disease—a double-edged sword. Neuron 35:419–432
Combarros O, Infante J, Llorca J, Berciano J (2003) Interleukin-1A (-889) genetic polymorphism increases the risk of multiple system atrophy. Mov Disord Off J Mov Disord Soc 18:1385–1386. doi:10.1002/mds.10540
Nishimura M, Kawakami H, Komure O et al (2002) Contribution of the interleukin-1beta gene polymorphism in multiple system atrophy. Mov Disord Off J Mov Disord Soc 17:808–811. doi:10.1002/mds.10124
Infante J, Llorca J, Berciano J, Combarros O (2005) Interleukin-8, intercellular adhesion molecule-1 and tumour necrosis factor-alpha gene polymorphisms and the risk for multiple system atrophy. J Neurol Sci 228:11–13. doi:10.1016/j.jns.2004.09.023
Furiya Y, Hirano M, Kurumatani N et al (2005) Alpha-1-antichymotrypsin gene polymorphism and susceptibility to multiple system atrophy (MSA). Brain Res Mol Brain Res 138:178–181. doi:10.1016/j.molbrainres.2005.04.011
Nishimura M, Kuno S, Kaji R, Kawakami H (2005) Influence of a tumor necrosis factor gene polymorphism in Japanese patients with multiple system atrophy. Neurosci Lett 374:218–221. doi:10.1016/j.neulet.2004.10.056
Shibao C, Garland EM, Gamboa A et al (2008) PRNP M129 V homozygosity in multiple system atrophy vs. Parkinson’s disease. Clin Auton Res Off J Clin Auton Res Soc 18:13–19. doi:10.1007/s10286-007-0447-7
Haïk S, Privat N, Adjou KT et al (2002) Alpha-synuclein-immunoreactive deposits in human and animal prion diseases. Acta Neuropathol (Berl) 103:516–520. doi:10.1007/s00401-001-0499-z
Jendroska K, Hoffmann O, Schelosky L et al (1994) Absence of disease related prion protein in neurodegenerative disorders presenting with Parkinson’s syndrome. J Neurol Neurosurg Psychiatry 57:1249–1251
Singleton AB, Farrer M, Johnson J et al (2003) alpha-Synuclein locus triplication causes Parkinson’s disease. Science 302:841. doi:10.1126/science.1090278
Farrer M, Kachergus J, Forno L et al (2004) Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol 55:174–179. doi:10.1002/ana.10846
Hernandez D, Paisan Ruiz C, Crawley A et al (2005) The dardarin G 2019 S mutation is a common cause of Parkinson’s disease but not other neurodegenerative diseases. Neurosci Lett 389:137–139. doi:10.1016/j.neulet.2005.07.044
Lincoln SJ, Ross OA, Milkovic NM et al (2007) Quantitative PCR-based screening of alpha-synuclein multiplication in multiple system atrophy. Parkinsonism Relat Disord 13:340–342. doi:10.1016/j.parkreldis.2006.12.005
Morris HR, Vaughan JR, Datta SR et al (2000) Multiple system atrophy/progressive supranuclear palsy: alpha-Synuclein, synphilin, tau, and APOE. Neurology 55:1918–1920
Ozawa T, Takano H, Onodera O et al (1999) No mutation in the entire coding region of the alpha-synuclein gene in pathologically confirmed cases of multiple system atrophy. Neurosci Lett 270:110–112
Ozawa T, Healy DG, Abou-Sleiman PM et al (2006) The alpha-synuclein gene in multiple system atrophy. J Neurol Neurosurg Psychiatry 77:464–467. doi:10.1136/jnnp.2005.073528
Ozawa T, Okuizumi K, Ikeuchi T et al (2001) Analysis of the expression level of alpha-synuclein mRNA using postmortem brain samples from pathologically confirmed cases of multiple system atrophy. Acta Neuropathol (Berl) 102:188–190
Vogt IR, Lees AJ, Evert BO et al (2006) Transcriptional changes in multiple system atrophy and Parkinson’s disease putamen. Exp Neurol 199:465–478. doi:10.1016/j.expneurol.2006.01.008
Langerveld AJ, Mihalko D, DeLong C et al (2007) Gene expression changes in postmortem tissue from the rostral pons of multiple system atrophy patients. Mov Disord Off J Mov Disord Soc 22:766–777. doi:10.1002/mds.21259
Ross OA, Vilariño-Güell C, Wszolek ZK et al (2010) Reply to: SNCA variants are associated with increased risk of multiple system atrophy. Ann Neurol 67:414–415. doi:10.1002/ana.21786
Al-Chalabi A, Dürr A, Wood NW et al (2009) Genetic variants of the alpha-synuclein gene SNCA are associated with multiple system atrophy. PLoS One 4:e7114. doi:10.1371/journal.pone.0007114
Simón-Sánchez J, Schulte C, Bras JM et al (2009) Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 41:1308–1312. doi:10.1038/ng.487
Satake W, Nakabayashi Y, Mizuta I et al (2009) Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet 41:1303–1307. doi:10.1038/ng.485
Yun JY, Lee W-W, Lee J-Y et al (2010) SNCA variants and multiple system atrophy. Ann Neurol 67:554–555. doi:10.1002/ana.21889
Guo XY, Chen YP, Song W et al (2014) SNCA variants rs2736990 and rs356220 as risk factors for Parkinson’s disease but not for amyotrophic lateral sclerosis and multiple system atrophy in a Chinese population. Neurobiol Aging. doi:10.1016/j.neurobiolaging.2014.07.014
Gan-Or Z, Bar-Shira A, Dahary D et al (2012) Association of sequence alterations in the putative promoter of RAB7L1 with a reduced parkinson disease risk. Arch Neurol 69:105–110. doi:10.1001/archneurol.2011.924
Guo X-Y, Chen Y-P, Song W et al (2014) An association analysis of the rs1572931 polymorphism of the RAB7L1 gene in Parkinson’s disease, amyotrophic lateral sclerosis and multiple system atrophy in China. Eur J Neurol Off J Eur Fed Neurol Soc 21:1337–1343. doi:10.1111/ene.12490
Vilariño-Güell C, Soto-Ortolaza AI, Rajput A et al (2011) MAPT H1 haplotype is a risk factor for essential tremor and multiple system atrophy. Neurology 76:670–672. doi:10.1212/WNL.0b013e31820c30c1
Wider C, Vilariño-Güell C, Jasinska-Myga B et al (2010) Association of the MAPT locus with Parkinson’s disease. Eur J Neurol Off J Eur Fed Neurol Soc 17:483–486. doi:10.1111/j.1468-1331.2009.02847.x
Sidransky E, Nalls MA, Aasly JO et al (2009) Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med 361:1651–1661. doi:10.1056/NEJMoa0901281
Srulijes K, Hauser A-K, Guella I et al (2013) No association of GBA mutations and multiple system atrophy. Eur J Neurol Off J Eur Fed Neurol Soc 20:e61–e62. doi:10.1111/ene.12086
Segarane B, Li A, Paudel R et al (2009) Glucocerebrosidase mutations in 108 neuropathologically confirmed cases of multiple system atrophy. Neurology 72:1185–1186. doi:10.1212/01.wnl.0000345356.40399.eb
Ozelius LJ, Foroud T, May S et al (2007) G2019S mutation in the leucine-rich repeat kinase 2 gene is not associated with multiple system atrophy. Mov Disord Off J Mov Disord Soc 22:546–549. doi:10.1002/mds.21343
Tan EK, Skipper L, Chua E et al (2006) Analysis of 14 LRRK2 mutations in Parkinson’s plus syndromes and late-onset Parkinson’s disease. Mov Disord Off J Mov Disord Soc 21:997–1001. doi:10.1002/mds.20875
Heckman MG, Schottlaender L, Soto-Ortolaza AI et al (2014) LRRK2 exonic variants and risk of multiple system atrophy. Neurology 83:2256–2261. doi:10.1212/WNL.0000000000001078
Hatano T, Kubo S, Sato S, Hattori N (2009) Pathogenesis of familial Parkinson’s disease: new insights based on monogenic forms of Parkinson’s disease. J Neurochem 111:1075–1093. doi:10.1111/j.1471-4159.2009.06403.x
Brooks JA, Houlden H, Melchers A et al (2011) Mutational analysis of parkin and PINK1 in multiple system atrophy. Neurobiol Aging 32(548):e5–e7. doi:10.1016/j.neurobiolaging.2009.11.020
Buervenich S, Sydow O, Carmine A et al (2000) Alcohol dehydrogenase alleles in Parkinson’s disease. Mov Disord Off J Mov Disord Soc 15:813–818
Buervenich S, Carmine A, Galter D et al (2005) A rare truncating mutation in ADH1C (G78Stop) shows significant association with Parkinson disease in a large international sample. Arch Neurol 62:74–78. doi:10.1001/archneur.62.1.74
Healy DG, Abou-Sleiman PM, Wood NW (2004) Genetic causes of Parkinson’s disease: UCHL-1. Cell Tissue Res 318:189–194. doi:10.1007/s00441-004-0917-3
Kim HS, Lee MS (2003) Frequencies of single nucleotide polymorphism in alcohol dehydrogenase7 gene in patients with multiple system atrophy and controls. Mov Disord Off J Mov Disord Soc 18:1065–1067. doi:10.1002/mds.10500
Healy DG, Abou-Sleiman PM, Quinn N et al (2005) UCHL-1 gene in multiple system atrophy: a haplotype tagging approach. Mov Disord Off J Mov Disord Soc 20:1338–1343. doi:10.1002/mds.20575
Renton AE, Majounie E, Waite A et al (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–268. doi:10.1016/j.neuron.2011.09.010
Goldman JS, Quinzii C, Dunning-Broadbent J et al (2014) Multiple system atrophy and amyotrophic lateral sclerosis in a family with hexanucleotide repeat expansions in C9orf72. JAMA Neurol 71:771–774. doi:10.1001/jamaneurol.2013.5762
Schottlaender LV, Holton JL, Houlden H (2014) Multiple system atrophy and repeat expansions in c9orf72. JAMA Neurol 71:1190–1191. doi:10.1001/jamaneurol.2014.1808
Scholz SW, Majounie E, Revesz T et al (2014) Multiple system atrophy is not caused by C9orf72 hexanucleotide repeat expansions. Neurobiol Aging. doi:10.1016/j.neurobiolaging.2014.08.033
Schottlaender L, Polke JM, Ling H et al (2014) The analysis of C9orf72 repeat expansions in a large series of clinically and pathologically diagnosed cases with atypical parkinsonism. Neurobiol Aging. doi:10.1016/j.neurobiolaging.2014.08.024
Chu K, Cho J-W, Song E-C, Jeon BS (2002) A patient with proximal myotonic myopathy and parkinsonism. Can J Neurol Sci J Can Sci Neurol 29:188–190
Celik Y, Turgut N, Balci K, Kabayel L (2006) Proximal myotonic dystrophy associated with parkinsonism. J Clin Neurosci Off J Neurosurg Soc Australas 13:275–276. doi:10.1016/j.jocn.2005.01.013
Annic A, Devos D, Destée A et al (2008) Early dopasensitive Parkinsonism related to myotonic dystrophy type 2. Mov Disord Off J Mov Disord Soc 23:2100–2101. doi:10.1002/mds.22239
Sansone V, Meola G, Perani D et al (2006) Glucose metabolism and dopamine PET correlates in a patient with myotonic dystrophy type 2 and parkinsonism. J Neurol Neurosurg Psychiatry 77:425–426. doi:10.1136/jnnp.2005.078451
Lim S-Y, Wadia P, Wenning GK, Lang AE (2009) Clinically probable multiple system atrophy with predominant parkinsonism associated with myotonic dystrophy type 2. Mov Disord Off J Mov Disord Soc 24:1407–1409. doi:10.1002/mds.22625
Cho S, Kim C-H, Cubells JF et al (2003) Variations in the dopamine beta-hydroxylase gene are not associated with the autonomic disorders, pure autonomic failure, or multiple system atrophy. Am J Med Genet A 120A:234–236. doi:10.1002/ajmg.a.20194
Federoff M, Jimenez-Rolando B, Nalls MA, Singleton AB (2012) A large study reveals no association between APOE and Parkinson’s disease. Neurobiol Dis 46:389–392. doi:10.1016/j.nbd.2012.02.002
Cairns NJ, Atkinson PF, Kovács T et al (1997) Apolipoprotein E e4 allele frequency in patients with multiple system atrophy. Neurosci Lett 221:161–164
Multhammer M, Michels A, Zintl M et al (2014) A large ApoE ε4/ε4 homozygous cohort reveals no association with Parkinson’s disease. Acta Neurol Belg 114:25–31. doi:10.1007/s13760-013-0223-5
Berciano J, Ferrer I (2005) Glial cell cytoplasmic inclusions in SCA2 do not express alpha-synuclein. J Neurol 252:742–744. doi:10.1007/s00415-005-0747-6
Factor SA, Qian J, Lava NS et al (2005) False-positive SCA8 gene test in a patient with pathologically proven multiple system atrophy. Ann Neurol 57:462–463. doi:10.1002/ana.20389
Kamm C, Healy DG, Quinn NP et al (2005) The fragile X tremor ataxia syndrome in the differential diagnosis of multiple system atrophy: data from the EMSA Study Group. Brain J Neurol 128:1855–1860. doi:10.1093/brain/awh535
Naka H, Ohshita T, Murata Y et al (2002) Characteristic MRI findings in multiple system atrophy: comparison of the three subtypes. Neuroradiology 44:204–209
Hagerman PJ, Greco CM, Hagerman RJ (2003) A cerebellar tremor/ataxia syndrome among fragile X premutation carriers. Cytogenet Genome Res 100:206–212 72856
Garland EM, Vnencak-Jones CL, Biaggioni I et al (2004) Fragile X gene premutation in multiple system atrophy. J Neurol Sci 227:115–118. doi:10.1016/j.jns.2004.08.013
Yabe I, Soma H, Takei A et al (2004) No association between FMR1 premutations and multiple system atrophy. J Neurol 251:1411–1412. doi:10.1007/s00415-004-0546-5
Manolio TA, Collins FS, Cox NJ et al (2009) Finding the missing heritability of complex diseases. Nature 461:747–753. doi:10.1038/nature08494
Sasaki H, Emi M, Iijima H et al (2011) Copy number loss of (src homology 2 domain containing)-transforming protein 2 (SHC2) gene: discordant loss in monozygotic twins and frequent loss in patients with multiple system atrophy. Mol Brain 4:24. doi:10.1186/1756-6606-4-24
Ferguson MC, Garland EM, Hedges L et al (2014) SHC2 gene copy number in multiple system atrophy (MSA). Clin Auton Res Off J Clin Auton Res Soc 24:25–30. doi:10.1007/s10286-013-0216-8
Sharp AJ, Locke DP, McGrath SD et al (2005) Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77:78–88. doi:10.1086/431652
Henrichsen CN, Vinckenbosch N, Zöllner S et al (2009) Segmental copy number variation shapes tissue transcriptomes. Nat Genet 41:424–429. doi:10.1038/ng.345
Bruder CEG, Piotrowski A, Gijsbers AACJ et al (2008) Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet 82:763–771. doi:10.1016/j.ajhg.2007.12.011
Stefanova N, Reindl M, Neumann M et al (2007) Microglial activation mediates neurodegeneration related to oligodendroglial alpha-synucleinopathy: implications for multiple system atrophy. Mov Disord Off J Mov Disord Soc 22:2196–2203. doi:10.1002/mds.21671
Block ML, Hong J-S (2007) Chronic microglial activation and progressive dopaminergic neurotoxicity. Biochem Soc Trans 35:1127–1132. doi:10.1042/BST0351127
Brand A, Bauer NG, Hallott A et al (2010) Membrane lipid modification by polyunsaturated fatty acids sensitizes oligodendroglial OLN-93 cells against oxidative stress and promotes up-regulation of heme oxygenase-1 (HSP32). J Neurochem 113:465–476. doi:10.1111/j.1471-4159.2010.06611.x
Stefanova N, Georgievska B, Eriksson H et al (2012) Myeloperoxidase inhibition ameliorates multiple system atrophy-like degeneration in a transgenic mouse model. Neurotox Res 21:393–404. doi:10.1007/s12640-011-9294-3
Bukhatwa S, Zeng B-Y, Rose S, Jenner P (2010) A comparison of changes in proteasomal subunit expression in the substantia nigra in Parkinson’s disease, multiple system atrophy and progressive supranuclear palsy. Brain Res 1326:174–183. doi:10.1016/j.brainres.2010.02.045
Schwarz L, Goldbaum O, Bergmann M et al (2012) Involvement of macroautophagy in multiple system atrophy and protein aggregate formation in oligodendrocytes. J Mol Neurosci MN 47:256–266. doi:10.1007/s12031-012-9733-5
Korolchuk VI, Menzies FM, Rubinsztein DC (2010) Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584:1393–1398. doi:10.1016/j.febslet.2009.12.047
Ebrahimi-Fakhari D, Cantuti-Castelvetri I, Fan Z et al (2011) Distinct roles in vivo for the ubiquitin-proteasome system and the autophagy-lysosomal pathway in the degradation of α-synuclein. J Neurosci Off J Soc Neurosci 31:14508–14520. doi:10.1523/JNEUROSCI.1560-11.2011
Rubinsztein DC, DiFiglia M, Heintz N et al (2005) Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 1:11–22
Stefanova N, Kaufmann WA, Humpel C et al (2012) Systemic proteasome inhibition triggers neurodegeneration in a transgenic mouse model expressing human α-synuclein under oligodendrocyte promoter: implications for multiple system atrophy. Acta Neuropathol (Berl) 124:51–65. doi:10.1007/s00401-012-0977-5
Wong MB, Goodwin J, Norazit A et al (2013) SUMO-1 is associated with a subset of lysosomes in glial protein aggregate diseases. Neurotox Res 23:1–21. doi:10.1007/s12640-012-9358-z
Höglinger GU, Melhem NM, Dickson DW et al (2011) Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet 43:699–705. doi:10.1038/ng.859
Welter D, Macarthur J, Morales J et al (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42:D1001–D1006. doi:10.1093/nar/gkt1229
Keller MF, Saad M, Bras J et al (2012) Using genome-wide complex trait analysis to quantify “missing heritability” in Parkinson’s disease. Hum Mol Genet 21:4996–5009. doi:10.1093/hmg/dds335
Lupski JR, Gonzaga-Jauregui C, Yang Y et al (2013) Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med 5:57. doi:10.1186/gm461
Lieber DS, Vafai SB, Horton LC et al (2012) Atypical case of Wolfram syndrome revealed through targeted exome sequencing in a patient with suspected mitochondrial disease. BMC Med Genet 13:3. doi:10.1186/1471-2350-13-3
Ionita-Laza I, Makarov V, Yoon S et al (2011) Finding disease variants in Mendelian disorders by using sequence data: methods and applications. Am J Hum Genet 89:701–712. doi:10.1016/j.ajhg.2011.11.003
Gonzaga-Jauregui C, Lupski JR, Gibbs RA (2012) Human genome sequencing in health and disease. Annu Rev Med 63:35–61. doi:10.1146/annurev-med-051010-162644
Guerreiro R, Wojtas A, Bras J et al (2013) TREM2 variants in Alzheimer’s disease. N Engl J Med 368:117–127. doi:10.1056/NEJMoa1211851
Schork NJ, Murray SS, Frazer KA, Topol EJ (2009) Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev 19:212–219. doi:10.1016/j.gde.2009.04.010
Koboldt DC, Steinberg KM, Larson DE et al (2013) The next-generation sequencing revolution and its impact on genomics. Cell 155:27–38. doi:10.1016/j.cell.2013.09.006
Green RC, Berg JS, Grody WW et al (2013) ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med Off J Am Coll Med Genet 15:565–574. doi:10.1038/gim.2013.73
Van El CG, Cornel MC, Borry P et al (2013) Whole-genome sequencing in health care: recommendations of the European Society of Human Genetics. Eur J Hum Genet EJHG 21:580–584. doi:10.1038/ejhg.2013.46
Acknowledgments
The authors work is supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services; project ZO1 AG000958.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Panel 1: Key technological and analytical tools used in human genetics
GENOME-WIDE ASSOCIATION (GWA) |
Definition: A GWA study uses extremely dense genome-wide genotyping to identify associations between genetic loci and the presence or absence of a trait. The goal is to identify genomic regions that contain risk alleles and this is done in a largely unbiased manner (i.e., without consideration of gene function or position). This is typically accomplished using millions of common genetic variants genotyped in large series of cases and controls. |
Application: Used primarily in the identification of risk loci for diseases and traits, it is largely limited to the identification of common risk alleles and explicitly tests the common disease common variant hypothesis. |
Limitations: In general GWA requires many thousands of cases and controls to reliably detect effects. Independent replication of identified loci is required. Does not reliably detect rare risk alleles. Identifies genetic regions that contain risk alleles, but does not identify either the causal risk allele or the affected gene. |
Cost and Use: Current cost ~ $200 per sample. Since initial use in 2005 GWA has been widely adopted and is still used extensively today. |
LINKAGE AND POSITIONAL CLONING |
Definition: Linkage and positional cloning served as the critical methods in the identification of mutations that caused single-gene (monogenic) disorders. Traditionally linkage analysis was performed in families with an obviously inherited disease. Polymorphic (variable) markers were run throughout the genomes of member of the family to identify regions of the genome that segregated with disease. The inference from this result was that a disease-segregating region was likely to contain the disease-causing mutation. Linkage was performed using 200-800 polymorphic markers spaced throughout the genome, although more recently this has been replaced by the use of SNP panels of hundreds of thousands of variants. Following the identification of positive linkage gene candidates within that region were sequenced to identify the causal mutation (this portion of the experiment was termed positional cloning). |
Application: Used in the identification of disease-causing mutations in highly informative (but usually rare) families |
Limitations: These methods were quite slow, with successful linkage and positional cloning projects often taking years. In general, families that were informative enough for this method are extremely rare, and in particular for a late-onset disease, challenging to collect (because multiple generations are required). |
Cost and Use: Relatively inexpensive, however, these methods have been largely supplanted by the use of exome sequencing, which combines elements of linkage and sequencing |
SECOND GENERATION SEQUENCING |
Definition: Second-generation sequencing (SGS) represents a major advance in molecular genetics. This method allows the generation of extremely large amounts of DNA sequence data, including the routine sequencing of human genomes. Most commonly thus far in human genetics, this method has been used in the context of exome sequencing. This involves sequencing of the protein-coding regions of the human genome. |
Application: The principal application has been in the identification of disease-causing mutations; exome sequencing allows an investigator to identify rare disease-segregating mutations rapidly and quite efficiently. More recently there has been interest in applying this method to large groups rather than families in an attempt to identify risk alleles. |
Limitations: The current methods are not able to easily detect certain types of variability (such as repeat expansions), and sequencing of certain parts of the genome (such as copy number variants) is unreliable. |
Cost and Use: Within a research setting exome sequencing costs approximately $500 per sample and whole genome sequencing $1,500 per sample; however, the price continues to decrease. Exome sequencing is widely used in genetics laboratories, but will likely be replaced by whole genome sequencing in short order. |
Use of exome/genome sequencing in a clinical setting: This is becoming a more cost-effective approach toward complex neurological diseases. However, there are mixed opinions on reporting of mutations in genes that were not intended to be the target. For example, finding a BRCA mutation in a patient being investigated because of a neurological disease. Guidelines have been developed [149, 150] to aid clinicians and laboratories, but this is a complicated matter and there is a large debate on clinical proceedings, ethical issues and consent. |
Rights and permissions
About this article
Cite this article
Federoff, M., Schottlaender, L.V., Houlden, H. et al. Multiple system atrophy: the application of genetics in understanding etiology. Clin Auton Res 25, 19–36 (2015). https://doi.org/10.1007/s10286-014-0267-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10286-014-0267-5