Abstract
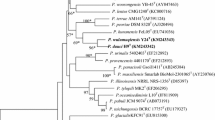
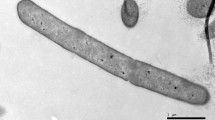
A Gram-staining positive, facultative aerobic bacterium, designated strain RH-N24T, was isolated from naked barley in South Korea. Cells of the isolate were observed to be motile rods by means of peritrichous flagella and showed catalase-positive and oxidase-negative reactions. Growth of strain RH-N24T was observed at 4–40 °C (optimum: 35–37 °C) and at pH 5.0–9.0 (optimum: pH 6.0–7.0). Chemotaxonomic data (major isoprenoid quinone: MK-7; DNA G + C content: 53.5 mol %; cell wall type: A1γ-meso-diaminopimelic acid; major fatty acids: anteiso-CB15:0 and CB16:0B) supported the affiliation of the isolate to the genus Paenibacillus. The major cellular polar lipids were identified as phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol and two unidentified polar lipids. Phylogenetic analysis based on 16S rRNA gene sequences also supported the conclusion that strain RH-N24T belonged to the genus Paenibacillus. Comparative 16S rRNA gene sequence analysis showed that strain RH-N24T was most closely related to Paenibacillus hunanensis FeL05T and Paenibacillus illinoisensis NRRL NRS-1356T with similarities of 94.64 and 94.54 %, respectively. On the basis of phenotypic and molecular properties, strain RH-N24T represents a novel species within the genus Paenibacillus for which the name Paenibacillus hordei sp. nov. is proposed. The type strain is RH-N24T (=KACC 15511T = JCM 17570T).


Similar content being viewed by others
References
Ash C, Priest FG, Collins MD (1993) Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 64:253–260
Chou JH, Chou YJ, Lin KY, Sheu SY, Sheu DS, Arun AB, Young CC, Chen WM (2007) Paenibacillus fonticola sp. nov., isolated from a warm spring. Int J Syst Evol Microbiol 57:1346–1350
Felsenstein J (2002) PHYLIP (phylogeny inference package), version 3.6a, Seattle: Department of Genetics, University of Washington, Seattle, WA, USA
Gomori G (1955) Preparation of buffers for use in enzyme studies. In: Colowick SP, Kaplan NO (eds) Methods in enzymology, vol 1. Academic Press, New York, pp 138–146
Gonzalez JM, Saiz-Jimenez C (2002) A fluorimetric method for the estimation of G + C mol % content in microorganisms by thermal denaturation temperature. Environ Microbiol 4:770–773
Jeon CO, Park W, Ghiorse WC, Madsen EL (2004) Polaromonas naphthalenivorans sp. nov., a naphthalene-degrading bacterium from naphthalene-contaminated sediment. Int J Syst Evol Microbiol 54:93–97
Jeon CO, Lim JM, Lee SS, Chung BS, Park DJ, Xu LH, Jiang CL, Kim CJ (2009) Paenibacillus harenae sp. nov., isolated from desert sand in China. Int J Syst Evol Microbiol 59:13–17
Jin HJ, Lv J, Chen SF (2011a) Paenibacillus sophorae sp. nov., a nitrogen-fixing species isolated from the rhizosphere of Sophora japonica. Int J Syst Evol Microbiol 61:767–771
Jin HJ, Zhou YG, Liu HC, Chen SF (2011b) Paenibacillus jilunlii sp. nov., a nitrogen-fixing species isolated from the rhizosphere of Begonia semperflorens. Int J Syst Evol Microbiol 61:1350–1355
Jung JY, Kim JM, Jin HM, Kim SY, Park W, Jeon CO (2011) Litorimonas taeanensis gen. nov., sp. nov., isolated from a sandy beach. Int J Syst Evol Microbiol 61:1534–1538
Khianngam S, Tanasupawat S, Akaracharanya A, Kim KK, Lee KC, Lee JS (2011) Paenibacillus xylanisolvens sp. nov., a xylan-degrading bacterium from soil. Int J Syst Evol Microbiol 61:160–164
Kim JM, Lee HJ, Kim SY, Song JJ, Park W, Jeon CO (2010) Analysis of fine-scale population structure of “Candidatus Accumulibacter phosphatis” using fluorescence in situ hybridization and flow cytometric sorting in enhanced biological phosphorus removal sludge. Appl Environ Microbiol 76:3825–3835
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kishore KH, Begum Z, Pathan AAK, Shivaji S (2010) Paenibacillus glacialis sp. nov., isolated from the Kafni glacier of the Himalayas, India. Int J Syst Evol Microbiol 60:1909–1913
Komagata K, Suzuki K (1988) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Lee SH, Park MS, Jung JY, Jeon CO (2012) Leuconostoc miyukkimchii sp. nov., isolated from brown algae (Undaria pinnatifida) kimchi. Int J Syst Evol Microbiol 62:1098–1103
Lim JM, Jeon CO, Lee JC, Xu LH, Jiang CL, Kim CJ (2006) Paenibacillus gansuensis sp. nov., isolated from desert soil of Gansu Province in China. Int J Syst Evol Microbiol 56:2131–2134
Liu Y, Liu L, Qiu F, Schumann P, Shi Y, Zou Y, Zhang X, Song W (2010) Paenibacillus hunanensis sp. nov., isolated from rice seeds. Int J Syst Evol Microbiol 60:1266–1270
Logan NA, Berge O, Bishop AH, Busse H-J, De Vos P, Fritze D, Heyndrickx M, Kämpfer P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
Lu S, Park M, Ro H-S, Lee DS, Park W, Jeon CO (2006) Analysis of microbial communities using culture-dependent and culture-independent approaches in an anaerobic/aerobic SBR reactor. J Microbiol 44:155–161
Minnikin DE, Patel PV, Alshamaony L, Goodfellow M (1977) Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Bacteriol 27:104–117
Montes MJ, Mercade E, Bozal N, Guinea J (2004) Paenibacillus antarcticus sp. nov., a novel psychrotolerant organism from the Antarctic environment. Int J Syst Evol Microbiol 54:1521–1526
Moon JC, Jung YJ, Jung JH, Jung HS, Cheong YR, Jeon CO, Lee KO, Lee SY (2011) Paenibacillus sacheonensis sp. nov., a xylanolytic and cellulolytic bacterium isolated from tidal flat sediment in Sacheon Bay, Korea. Int J Syst Evol Microbiol 61:2753–2757
Park MH, Traiwan J, Jung MY, Nam YS, Jeong JH, Kim W (2011) Paenibacillus chungangensis sp. nov., isolated from a tidal-flat sediment. Int J Syst Evol Microbiol 61:281–285
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acid Res 35:7188–7196
Rivas R, Gutierrez C, Abril A, Mateos PF, Martinez-Molina E, Ventosa A, Velazquez E (2005a) Paenibacillus rhizosphaerae sp. nov., isolated from the rhizosphere of Cicer arietinum. Int J Syst Evol Microbiol 55:1305–1309
Rivas R, Mateos PF, Martínez-Molina E, Velázquez E (2005b) Paenibacillus xylanilyticus sp. nov., an airborne xylanolytic bacterium. Int J Syst Evol Microbiol 55:405–408
Rodríguez-Díaz M, Lebbe L, Rodelas B, Heyrman J, De Vos P, Logan NA (2005) Paenibacillus wynnii sp. nov., a novel species harbouring the nifH gene, isolated from Alexander Island, Antarctica. Int J Syst Evol Microbiol 55:2093–2099
Rosselló-Mora R, Amann R (2001) The species concept for prokaryotes. FEMS Microbiol Rev 25:39–67
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. Newark, DE: MIDI Inc
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477
Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K (1997a) Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol 47:289–298
Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K (1997b) Emended description of Paenibacillus amylolyticus and description of Paenibacillus illinoisensis sp. nov. Int J Syst Bacteriol 47:299–306
Šmerda J, Sedláček I, Páčová Z, Durnová E, Smíšková A, Havel L (2005) Paenibacillus mendelii sp. nov., from surface-sterilized seeds of Pisum sativum L. Int J Syst Evol Microbiol 55:2351–2354
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P (ed) Methods for general and molecular bacteriology. American Society for Microbiology, Washington D.C., pp 607–654
Tang QY, Yang N, Wang J, Xie YQ, Ren B, Zhou YG, Gu MY, Mao J, Li WJ, Shi YH, Zhang LX (2011) Paenibacillus algorifonticola sp. nov., isolated from a cold spring. Int J Syst Evol Microbiol 61:2167–2172
Wang L, Baek SH, Cui Y, Lee HG, Lee ST (2012) Paenibacillus sediminis sp. nov., a xylanolytic bacterium isolated from a tidal flat. Int J Syst Evol Microbiol 62:1284–1288
Yoon JH, Kang SJ, Yeo SH, Oh TK (2005) Paenibacillus alkaliterrae sp. nov., isolated from an alkaline soil in Korea. Int J Syst Evol Microbiol 55:2339–2344
Acknowledgments
These efforts were supported by the Technology Development Program for Agriculture and Forestry (TDPAF) of the Ministry for Agriculture, Forestry and Fisheries and the project on survey and excavation of Korean indigenous species of the National Institute of Biological Resources (NIBR) under the Ministry of Environment, Republic of Korea. The GenBank accession number for the 16S rRNA gene sequence of strain RH-N24T is HQ833590.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, J.M., Lee, S.H., Lee, S.H. et al. Paenibacillus hordei sp. nov., isolated from naked barley in Korea. Antonie van Leeuwenhoek 103, 3–9 (2013). https://doi.org/10.1007/s10482-012-9775-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-012-9775-2