Abstract
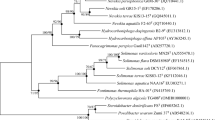
A strictly aerobic, Gram-stain-negative, rod-shaped and red-orange pigmented bacterium, designated strain KMM 6395T, was isolated from the green alga Cladophora stimpsoni and subjected to a polyphasic taxonomic study. Phylogenetic analysis based on 16S rRNA gene sequencing revealed that the novel strain affiliated to the family Hyphomonadaceae of the class Alphaproteobacteria being most closely related to the type strain of the single species of the genus Litorimonas, Litorimonas taeanensis G5T, with 16S rRNA gene sequence similarity of 96.8 %. Strain KMM 6395T grew with 1–5 % NaCl and at 4–35 °C, hydrolysed starch and Tween 80. The DNA G+C content was 48.7 mol%. The prevalent fatty acids were C18:1 ω7c, C19:1 ω8c and C18:1 ω7c 10-methyl. The polar lipid profile was characterized by the presence of phosphatidylglycerol, monoglycosyldiglyceride, glucuronopyranosyldiglyceride and an unidentified glycolipid. The major respiratory quinone was Q-10. The significant molecular distinctiveness between the novel isolate and its nearest neighbour, L. taeanensis G5T, were strongly supported by the differences in physiological and biochemical tests. Therefore, strain KMM 6395T represents a novel species of the genus Litorimonas, for which the name Litorimonas cladophorae sp. nov. is proposed. The type strain is KMM 6395T (=KCTC 23968T = LMG 26985T). The emended descriptions of the genus Litorimonas and L. taeaensis are also provided.


Similar content being viewed by others
References
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Collins MD, Shah HM (1984) Fatty acid, menaquinone and polar lipid composition of Rothia dentosacariosa. Arch Microbiol 137:247–249
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gonzalez JM, Saiz-Jimenez C (2002) A fluorimetric method for the estimation of G+C mol% content in microorganisms by thermal denaturation temperature. Environ Microbiol 4:770–773
Jung JY, Kim JM, Jin HM, Kim SY, Park W, Jeon CO (2011) Litorimonas taeanensis gen. nov., sp. nov., isolated from a sandy beach. Int J Syst Evol Microbiol 61:1534–1538
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Komagata K, Suzuki K-I (1987) Lipid and cell wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Lemos ML, Toranzo AE, Barja JL (1985) Modified medium for oxidation-fermentation test in the identification of marine bacteria. Appl Environ Microbiol 40:1541–1543
Nedashkovskaya OI, Kim SB, Han SK, Rhee MS, Lysenko AM, Falsen E, Frolova GM, Mikhailov VV, Bae KS (2004) Ulvibacter litoralis gen. nov., sp. nov., a novel member of the family Flavobacteriaceae isolated from the green alga Ulva fenestrata. Int J Syst Evol Microbiol 54:119–123
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Rzhetsky A, Nei M (1992) A simple method for estimating and testing minimum evolution trees. Mol Biol Evol 9:945–967
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russell DW (2001) Molecular Cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20:16
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Acknowledgments
We thank Dr. Ji Young Jung (Chung-Ang University, Seoul, Republic of Korea) for providing us with the type strain L. taenensis G5T. Dr. D.V. Fomin (Cooperative Far Eastern Center of Electron Microscopy, Vladivostok, Russia) is gratefully acknowledged for his excellent technical assistance. This research was supported by grants of the Presidium of the Russian Academy of Sciences “Molecular and Cell Biology”, the Presidium of the Far-Eastern Branch of the Russian Academy of Sciences no. 12-III-A-06-105, the government of Russian Federation for the state support of scientific investigations conducting under the guidance of the leading researchers at the Russian education institutions of the high professional education, agreement no. 11.G34.31.0010 and the Russian Foundation for Basic Research (RFBR) no. 11-04-00781.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nedashkovskaya, O.I., Kukhlevskiy, A.D., Zhukova, N.V. et al. Litorimonas cladophorae sp. nov., a new alphaproteobacterium isolated from the Pacific green alga Cladophora stimpsoni, and emended descriptions of the genus Litorimonas and Litorimonas taeaensis . Antonie van Leeuwenhoek 103, 1263–1269 (2013). https://doi.org/10.1007/s10482-013-9906-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-9906-4