Abstract
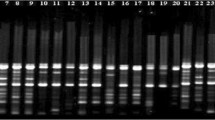
Estimation of variability and genetic relationships among breeding materials is one of the important strategies in crop improvement programs. Morphological (plant height, spike length, a number of florets/spike), physiological (chlorophyll content, chlorophyll fluorescence, and rapid light curve parameters) and Directed amplification of minisatellite DNA (DAMD) markers were used to investigate the relationships among 50 Gladiolus cultivars. Cluster analysis based on morphological data, physiological characteristics, molecular markers, and cumulative data discriminated all cultivars into seven, five, seven, and six clusters in the unweighted pair-group method using arithmetic mean (UPGMA) dendrogram, respectively. The results of the principal coordinate analysis (PCoA) also supported UPGMA clustering. Variations among the Gladiolus cultivars at phenotypic level could be due to the changes in physiology, environmental conditions, and genetic variability. DAMD analysis using 10 primers produced 120 polymorphic bands with 80% polymorphism showing polymorphic information content (PIC = 0.28), Marker index (MI = 3.37), Nei’s gene diversity (h = 0.267), and Shannon’s information index (I = 0.407). Plant height showed a positive significant correlation with Spike length and Number of florets/spike (r = 0.729, p < 0.001 and r = 0.448, p = 0.001 respectively). Whereas, Spike length showed positive significant correlation with Number of florets/spike (r = 0.688, p < 0.001) and Chlorophyll content showed positive significant correlation with Electron transport rate (r = 0.863, p < 0.001). Based on significant morphological variations, high physiological performance, high genetic variability, and genetic distances between cultivars, we have been able to identify diverse cultivars of Gladiolus that could be the potential source as breeding material for further genetic improvement in this ornamental crop.







Similar content being viewed by others
References
Alvino A, Andria RD, Cerio L, Mori M, Sorrentino G (1998) Physiological response of Gladiolus to carbonated water application under two water regimes. Adv Hortic Sci 12:145–152
Archana B, Patil AA, Hunje R, Patil VS (2008) Studies on genetic variability analysis in Gladiolus hybrids. J Ornam Hortic 11(2):121–126
Arora JS, Khanna K (1991) Genotypic and environmental interaction of some quantitative traits in Gladiolus. Indian J Hortic 48(4):86–88
Bachmann K (1992) Phenotypic similarity and genetic relationship among populations of Microseris bigelovii (Asteraceae: Lactuceae). Bot Acta 105:337–342
Beer SB, Vilenkin A, Weil M, Veste L, Eshel S (1998) Measuring photosynthetic rates in seagrasses by pulse amplitude modulated (PAM) fluorometry. Mar Ecol Prog Ser 174:293–300
Cerovic ZG, Masdoumier G, Ghozlen NB, Latouche G (2012) A new optical leaf-clip meter for simultaneous non-destructive assessment of leaf chlorophyll and epidermal flavonoids. Physiol Plant 146:251–260
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13
Goldblatt P, Manning J (1998) Gladiolus in Southern Africa. Timber Press, Portland
Goldblatt P, Manning JC (2008) Systematics of the southern African genus Ixia (Iridaceae). 1. The I. rapunculoides complex. Bothalia 38:1–23
Goldblatt P, Manning JC, Bernhardt P (2001) Radiation of pollination systems in Gladiolus (Iridaceae: Crocoideae) in southern Africa. Ann Mo Bot Gard 88:713–734
Hossain MD, Talukder KH, Asaduzzaman M, Mahmud F, Amin N, Sayed MA (2011) Study on morphological characteristics of different genotypes of Gladiolus flower. J Sci Found 9:1–8
Hu JB, Li JW, Wang LJ, Liu LJ, Si SW (2011) Utilization of a set of high polymorphism DAMD markers for genetic analysis of a cucumber germplasm collection. Acta Physiol Plant 33:227–231
Irawan B, Gruber F, Finkeldey R, Gailing O (2016) Linking indigenous knowledge, plant morphology, and molecular differentiation: the case of ironwood (Eusideroxylon zwageri Teijsm. et Binn.). Genet Resour Crop Evol 63:1297–1306
Jingang W, Hongkun J, Shufang G, Daidi C (2006) RAPD Analysis of 12 general species of Gladiolus hybridus Hort. J Northeast Agric Univ 13:112–115
Jingang W, Ying G, Daidi C, Shenkui L, Chuanpin Y (2008) ISSR analysis of 26 general species of Gladiolus hybridus Hort. J Northeast Agric Univ 15(4):6–10
Kamble BS, Reddy BS, Patil RT, Kulkarni BS (2004) Performance of Gladiolus (Gladiolus hybridus hort) cultivars for flowering and flower quality. J Ornam Hortic 7:51–56
Kumar PN, Raju DVS (2007) Dormancy in Gladiolus: the cause and remedy—a review. Agric Rev 28(4):309–312
Kumar R, Yadav DS (2005) Evaluation of Gladiolus cultivars under sub-tropical hill of Meghalaya. J Ornam Hortic 8(2):86–90
Lal SD, Pant CC (1989) Some newly developed hybrids of Gladiolus. Prog Hort 21:189–193
Lal SD, Shah A, Seth JN (1985) Genetic variability in Gladiolus II. Correlation between growth and yield contributing characters. Prog Hortic 17(1):31–34
Malik K, Krishan Pal (2014) Genetic divergence and relationship analysis among twenty two populations of Gladiolus cultivars by morphological and RAPD PCR tool. Int J Educ Sci Res Rev 1(6):1–8
Maxwell K, Johnson GN (2000) Chlorophyll fluorescence- a practical guide. J Exp Bot 51:659–668
Mukhopadhyay A, Banker GZ (1987) Gladiolus flower round the year Bangalore region. Indian Hortic 31(4):19–20
Nahed GAA, Lobna ST, Soad MMI (2009) Some studies on the effect of putrescine, ascorbic acid and thiamine on growth, flowering and some chemical constituents of Gladiolus plants at Nubaria. Oze J App Sci 2:169–179
Nasir IA, Jamal A, Rahman Z, Husnain T (2012) Molecular analyses of Gladiolus lines with improved resistance against fusarium wilt. Pak J Bot 44:73–79
Neeraj HP, Jha PB (2000) Evaluation of Gladiolus germplasm under North Bihar conditions. Indian J Hortic 57:178–181
Pant CC, Lal SD (1991) Correlation studies in Gladiolus. Prog Hortic 24:6–8
Parker PG, Snow AA, Sehug MD, Booton GC, Fuerst PA (1998) What molecular can tell us about populations: choosing and using a molecular marker. Ecology 79:361–382
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Persson HA, Runpeenen K, Mollerstedt LK (2000) Identification of culinary rhubarb (Rheum spp.) cultivars using morphological characterization and RAPD markers. J Hortic Sci Biotechnol 75:684–689
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Pragya Bhat KV, Misra RL, Ranjan JK (2010) Analysis of diversity and relationships among Gladiolus cultivars using morphological and RAPD markers. Ind J Agric Sci 80(9):766–772
Ranjan P, Bhat KV, Misra RL, Singh SK, Ranjan JK (2010) Genetic relationships of Gladiolus cultivars inferred from fluorescence based AFLP markers. Sci Hortic 123:562–567
Rascher U, Liebig M, Luttge U (2000) Evaluation of instant light-response curves of chlorophyll fluorescence parameters obtained with a portable chlorophyll fluorometer onsite in the field. Plant Cell Environ 23:1397–1405
Raycheva T, Stoyanov K, Denev I (2011) Genetic diversity and molecular taxonomy study of three genera from Iridaceae family in the Bulgarian flora based on ISSR markers. Biotechnol Biotechnol Equip 25:2484–2488
Rohlf FJ (2000) NTSYS-pc: Numerical taxonomy and multivariate analysis system, version 2.1, user guide. Exeter Software, New York
Schreiber U, Gademann R, Ralph PJ, Larkum AWD (1997) Assessment of the photosynthetic performance of Prochloron in Lissoclinum patella in hospital by chlorophyll fluorescence measurements. Plant Cell Physiol 38:945–951
Singh N, Pal AK, Roy RK, Tewari SK, Tamta S, Rana TS (2016) Assessment of genetic variation and population structure in indigenous Gladiolus cultivars inferred from molecular markers. The Nucleus 59:235–244
Singh N, Meena B, Pal AK, Roy RK, Tewari SK, Tamta S, Rana TS (2017a) Nucleotide diversity and phylogenetic relationships among Gladiolus cultivars and related taxa of family Iridaceae. J Genet 96:135–145
Singh N, Pal AK, Roy RK, Tewari SK, Tamta S, Rana TS (2017b) Development of cpSSR markers for analysis of genetic diversity in Gladiolus cultivars. Plant Gene 10:31–36
Swaroop K (2010) Morphological variation and evolution of Gladiolus germplasm. Indian J Agric Sci 80(8):78–81
Takatsu Y, Miyamoto M, Inoue E, Yamada Y, Manabe T, Kasumi M, Hayashi M, Sakuma F, Marubashi W, Niwa M (2001) Interspecific hybridization among wild Gladiolus Species of South Africa based on randomly amplified polymorphic DNA markers. Sci Hortic 91:339–348
Wang J, Bao MZ (2005) Characterization of genetic relationships in pansy (Viola wittrockiana) inbred lines using morphological traits and RAPD markers. J Hortic Sci Biotechnol 80:537–542
Wongchaochant S, Doi M, Imanishi H (2002) Phylogenetic classification of Freesia spp. by morphological and physiological characteristics and RAPD markers. J Japan Soc Hortic Sci 71:758–764
Yadav CB, Muthamilarasan M, Pandey G, Khan Y, Prasad M (2014) Development of novel microRNA-based genetic markers in foxtail millet for genotyping applications in related grass species. Mol Breed 34:2919–2924
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10528_2017_9835_MOESM1_ESM.pptx
Fig. S1 Two-dimensional plot of the first (87.62%) and second (11.72%) principal coordinate axes, derived from principal coordinate analysis of morphological data of 50 Gladiolus cultivars. (Numbers are equivalent to those listed in Table 1). Supplementary material 1 (PPTX 72 kb)
10528_2017_9835_MOESM2_ESM.pptx
Fig. S2 Two-dimensional plot of the first (98.74%) and second (0.90%) principal coordinate axes, derived from principal coordinate analysis of physiological data of 50 Gladiolus cultivars. (Numbers are equivalent to those listed in Table 1). Supplementary material 2 (PPTX 54 kb)
10528_2017_9835_MOESM3_ESM.pptx
Fig. S3 Two-dimensional plot of the first (8.59%) and second (7.30%) principal coordinate axes, derived from principal coordinate analysis of DAMD data of 50 Gladiolus cultivars. (Numbers are equivalent to those listed in Table 1). Supplementary material 3 (PPTX 70 kb)
Rights and permissions
About this article
Cite this article
Singh, N., Pal, A.K., Roy, R.K. et al. Characterization of Gladiolus Germplasm Using Morphological, Physiological, and Molecular Markers. Biochem Genet 56, 128–148 (2018). https://doi.org/10.1007/s10528-017-9835-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-017-9835-4