Abstract
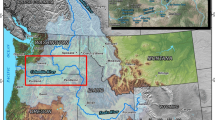
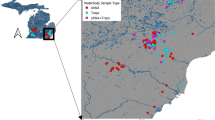
In light of the ongoing spread and adverse impacts of invasive species, there is an urgent need to develop more effective monitoring and management strategies. Such efforts are constrained by our limited capacity to efficiently detect invasive species. Here, we present the case of Bythotrephes longimanus (spiny water flea) invasion into Wisconsin lakes. Detecting Bythotrephes has proven to be challenging due to its capacity to persist at low densities and its highly seasonal population dynamics. We use Bythotrephes to explore detection using three monitoring methods: zooplankton net tows, environmental DNA (eDNA), and sampling of Bythotrephes tail spine subfossils in sediments. Detection probabilities were highly seasonal for both the net tow and eDNA sampling methods—though detections occurred one to two weeks earlier in net tows—and seasonal targeting substantially improved detection by both methods. Conversely, Bythotrephes spine subfossils were found in all 10 lakes with confirmed Bythotrephes populations and in all five samples taken from each lake, except for a single lake where four of the five samples had subfossils. This method was insensitive to seasonally varying population densities as sediments integrate over variation in population densities. In this case, detection and abundance estimation were well covered by sediments and zooplankton nets, respectively, and eDNA provided little additional benefit to surveillance. Our work highlights the importance of choosing methods that address both species life history and monitoring objectives when designing surveillance programs.



Similar content being viewed by others
References
Beranek A (2012) An assessment of the long-term phenology and impact of Bythotrephes longimanus in Island Lake Reservoir, Minnesota, using sediment records. University of Minnesota, Master’s Thesis
Berg D, Garton D (1988) Seasonal abundance of the exotic predatory cladoceran, Bythotrephes Cederstroemi, in Western Lake Erie. J Great Lakes Res 14:479–488
Bobeldyk AM, Rüegg J, Lamberti GA (2015) Freshwater hotspots of biological invasion are a function of species–pathway interactions. Hydrobiologia 746:363–373
Branstrator DK, Beranek AE, Brown ME, Hembre LK, Engstrom DR (2017) Colonization dynamics of the invasive predatory cladoceran, Bythotrephes longimanus, inferred from sediment records. Limnol Oceanogr 62:1096–1110
Brown ME, Branstrator DK (2011) Patterns in the abundance, phenology, and hatching of the resting egg stage of the invasive zooplankter Bythotrephes longimanus: implications for establishment. Biol Invasions 13:2547–2559
Churchill C (2013) Spatio-temporal spawning and larval dynamics of a zebra mussel (Dreissena polymorpha) population in a North Texas Reservoir: implications for invasions in the southern United States. Aquat Invasions 8:389–406
de Souza LS, Godwin JC, Renshaw MA, Larson E (2016) Environmental DNA (eDNA) detection probability is influenced by seasonal activity of organisms. PLoS ONE 11:e0165273
Deiner K, Altermatt F (2014) Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 9:e88786
Deiner K, Fronhofer E, Machler E, Walser J, Altermatt F (2016) Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat Commun 7:12544
Delaney DG, Leung B (2010) An empirical probability model of detecting species at low densities. Ecol Appl 20:1162–1172
Doi H, Uchii K, Takahara T, Matsuhashi S, Yamanaka H, Minamoto T (2015) Use of droplet digital PCR for estimation of fish abundance and biomass in environmental DNA surveys. PLoS ONE 10:e0122763
Drake JM (2004) Allee effects and the risk of biological invasion. Risk Anal 24(4):795–802
Ficetola GF, Miaud C, Pompanon F, Taberlet P (2008) Species detection using environmental DNA from water samples. Biol Lett 4:423–425
Gardham S, Hose G, Stephenson S, Chariton A (2014) DNA metabarcoding meets experimental ecotoxicology: advancing knowledge on the ecological effects of copper in freshwater ecosystems. Adv Ecol Res 51:79–104
Harvey CT, Qureshi SA, MacIsaac HJ (2009) Detection of a colonizing, aquatic, non-indigenous species. Divers Distrib 15:429–437
Hulme PE (2006) Beyond control: wider implications for the management of biological invasions. J Appl Ecol 43(5):835–847
Jerde CL, Mahon AR, Chadderton WL, Lodge DM (2011) “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv Lett 4:150–157
Kelly RP, Port JA, Yamahara KM, Crowder LB (2014) Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE 9:e86175
Kerfoot WC, Hobmeier MM, Yousef F, Lafrancois BM, Maki RP, Hirsch JK (2016) A plague of waterfleas (Bythotrephes): impacts on microcrustacean community structure, seasonal biomass, and secondary production in a large inland-lake complex. Biol Invasions 18:1121–1145
Kim N, Yan ND (2013) Food limitation impacts life history of the predatory cladoceran Bythotrephes longimanus, an invader to North America. Hydrobiologia 715:213–224
Leung B, Delaney DG (2006) Managing sparse data in biological invasions: a simulation study. Ecol Model 198:229–239
Lodge DM, Williams S, MacIsaac HJ, Hayes KR, Leung B, Reichard S, Mack RN, Moyle P, Smith M, Andow DA, Carlton JT, McMichael A (2006) Biological invasions: recommendations for U.S. policy and management. Ecol Appl 16(6):2035–2054
Mächler E, Deiner K, Steinmann P, Altermatt F (2014) Utility of environmental DNA for monitoring rare and indicator macroinvertebrate species. Freshw Sci 33:1174–1183
Magurran AE, McGill BJ (2011) Biological diversity: frontiers in measurement and assessment. Oxford University Press Inc., New York
Mahon AR, Jerde CL, Galaska M, Bergner JL, Chadderton WL, Lodge DM, Hunter ME, Nico LG (2013) Validation of eDNA surveillance sensitivity for detection of Asian carps in controlled and field experiments. PLoS ONE 8:e58316
Mwatawala MW, De Meyer M, Makundi RH, Maerere AP (2006) Seasonality and host utilization of the invasive fruit fly, Bactrocera invadens (Dipt., Tephritidae) in central Tanzania. J Appl Entomol 130:530–537
Nielsen AL, Hamilton GC, Shearer PW (2011) Seasonal phenology and monitoring of the non-native Halyomorpha halys (Hemiptera: Pentatomidae) in soybean. Environ Entomol 40:231–238
O’Connell AF, Talancy NW, Bailey LL, Sauer JR, Cook R, Gilbert AT (2006) Estimating site occupancy and detection probability parameters for meso- and large mammals in a coastal ecosystem. J Wildl Manag 70:1625–1633
Olson ZH, Briggler JT, Williams RN (2012) An eDNA approach to detect eastern hellbenders (Cryptobranchus a. alleganiensis) using samples of water. Wildl Res 39(7):629–636
Pierson TW, McKee AM, Spear SF, Maerz JC, Camp CD, Glenn TC (2016) Detection of an enigmatic Plethodontid salamander using environmental DNA. Copeia 104:78–82
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2014) Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol Ecol Resour 14:109–116
R Core Team (2016) R: A language and environment for statistical computing. R Foundation for Statistical Computing
Rees HC, Maddison BC, Middleditch DJ, Patmore JRM, Gough KC (2014) REVIEW: the detection of aquatic animal species using environmental DNA—a review of eDNA as a survey tool in ecology. J Appl Ecol 51:1450–1459
Renshaw MA, Olds BP, Jerde CL, McVeigh MM, Lodge DM (2015) The room temperature preservation of filtered environmental DNA samples and assimilation into a phenol–chloroform–isoamyl alcohol DNA extraction. Mol Ecol Resour 15:168–176
Rew LJ, Maxwell BD, Dougher FL, Aspinall R (2006) Searching for a needle in a Haystack: evaluating survey methods for non-indigenous plant species. Biol Invasions 8:523–539
Roussel J-M, Paillisson J-M, Tréguier A, Petit E (2015) The downside of eDNA as a survey tool in water bodies. J Appl Ecol 52:823–826
Simberloff D (2011) How common are invasion-induced ecosystem impacts? Biol Invasions 13:1255–1268
Simberloff D (2014) Biological invasions: What’s worth fighting and what can be won? Ecol Eng 65:112–121
Takahara T, Minamoto T, Yamanaka H, Doi H, Kawabata Z (2012) Estimation of fish biomass using environmental DNA. PLoS ONE 7:e35868
Thomsen PF, Kielgast J, Iversen LL, Wiuf C, Rasmussen M, Gilbert MTP, Orlando L, Willerslev E (2012) Monitoring endangered freshwater biodiversity using environmental DNA. Mol Ecol 21:2565–2573
Tréguier A, Paillisson J-M, Dejean T, Valentini A, Schlaepfer MA, Roussel J-M (2014) Environmental DNA surveillance for invertebrate species: advantages and technical limitations to detect invasive crayfish Procambarus clarkii in freshwater ponds. J Appl Ecol 51(4):871–879
Turner CR, Uy KL, Everhart RC (2015) Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol Cons 183:93–102
US-EPA (2016) United States Environmental Protection Agency: Zooplankton sampling methods
Vine SJ, Crowther MS, Lapidge SJ, Dickman CR, Mooney N, Piggott MP, English AW (2009) Comparison of methods to detect rare and cryptic species: a case study using the red fox (Vulpes vulpes). Wildl Res 36:436–446
Walsh JR, Carpenter SR, Vander Zanden MJ (2016a) Invasive species triggers a massive loss of ecosystem services through a trophic cascade. Proc Natl Acad Sci 113:4081–4085
Walsh JR, Munoz SE, Vander Zanden MJ (2016b) Outbreak of an undetected invasive species triggered by a climate anomaly. Ecosphere 7:e01628. https://doi.org/10.1002/ecs2.1628
Wittmann MJ, Lewis MA, Young JD, Yan ND (2011) Temperature-dependent Allee effects in a stage-structured model for Bythotrephes establishment. Biol Invasions 13:2477–2497
Yan N, Pawson TW (1997a) Changes in the crustacean zooplankton community of Harp Lake, Canada, following invasion by Bythotrephes cederstroemi. Freshw Biol 37:409–425
Yan ND, Pawson TW (1997b) Seasonal variation in the size and abundance of the invading Bythotrephes in Harp Lake, Ontario, Canada. Hydrobiologia 361:157–168
Yan ND, Leung B, Lewis MA, Peacor SD (2011) The spread, establishment and impacts of the spiny water flea, Bythotrephes longimanus, in temperate North America: a synopsis of the special issue. Biol Invasions 13:2423–2432
Young JD, Yan ND (2008) Modifications of the diel vertical migration of Bythotrephes longimanus by the cold-water zooplanktivore, Coregonus artedi. Freshw Biol 53(5):981–995
Young JD, Strecker AL, Yan ND (2011) Increased abundance of the non-indigenous zooplanktivore, Bythotrephes longimanus, is strongly correlated with greater spring prey availability in Canadian Shield lakes. Biol Invasions 13:2605–2619
Acknowledgements
We would like to thank Carol Warden for field and lab work conducted in Stormy Lake, Gile Flowage, and Lake Gogebic, the North Temperate Lakes Long Term Ecological Research program (NTL-LTER) field crew, including Pam Montz for field and lab work conducted in Trout Lake, and Ted Bier and Kirsten Rhude for field and lab work conducted in Lakes Mendota and Monona. We also thank Kassandra Ford, Marco Scarasso, Michaela Kromrey, Bridget Murphy, Sommer Kuhn, and Michael Josephson for their work in the lab and field. We thank Scott Van Egeren and Maureen Ferry for suggestions and advice in designing and applying this study. Finally, we thank multiple colleagues for comments and friendly review including Emily Stanley, Steve Carpenter, Tony Ives, and Randy Jackson. This work was funded by the Wisconsin Department of Natural Resources, the NSF NTL LTER Program (Grants DEB-0217533 and DEB-1440297), and the office of the UW-Madison Vice Chancellor for Research and Graduate Education.
Author information
Authors and Affiliations
Contributions
JRW, MJS, TPS, PJK, and MJVZ designed research; JRW, MJS, and TPS performed research; JRW, MJS, TPS, PJK, and MJVZ analyzed data; JRW, MJS, TPS, PJK, and MJVZ wrote the paper.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Walsh, J.R., Spear, M.J., Shannon, T.P. et al. Using eDNA, sediment subfossils, and zooplankton nets to detect invasive spiny water flea (Bythotrephes longimanus). Biol Invasions 21, 377–389 (2019). https://doi.org/10.1007/s10530-018-1862-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-018-1862-5