Abstract
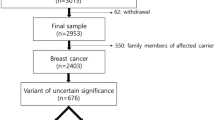
Characterizing the pathogenicity of BRCA1 variants of uncertain significance (VUSs) is a major bottleneck in clinical management of BRCA1-associated breast cancer. Saturation genome editing (SGE) was recently reported as an innovative laboratory-based approach to assess the pathogenicity of BRCA1 variants. We combined clinical phenotypes and SGE score to identify the pathogenicity of BRCA1 VUSs detected in a cohort of 8,085 breast cancer patients. According to SGE function score, 33 out of 144 BRCA1 VUSs detected were classified into “loss of function” (n = 13), “intermediate” (n = 2), and “functional” (n = 18) groups. Compared with non-carriers, “loss of function” VUS carriers (n = 19) presented significantly worse clinicopathological characteristics. These included younger age at breast cancer diagnosis (44.4 years vs. 51.2 years, P = 0.01), stronger family history of any cancer (57.9% vs. 32.3%, P = 0.017) especially breast or ovarian cancer (47.4% vs. 9.3%, P < 0.001), more bilateral breast cancer (31.6% vs. 3.4%, P < 0.001), and triple-negative breast cancer (47.4% vs. 12.8%, P < 0.001), which were comparable to those of pathogenic variant carriers. In contrast, the clinical phenotypes of “functional” VUS carriers were similar to those of non-carriers. These results indicated that SGE was a reliable method in BRCA1 variant classification. Combining SGE function score and the available evidence, twelve out of 33 BRCA1 VUSs were reclassified as pathogenic or likely pathogenic variants and one was benign.



Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72(5):1117–1130. https://doi.org/10.1086/375033
King MC, Marks JH, Mandell JB, New York Breast Cancer Study G (2003) Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302(5645):643–646. https://doi.org/10.1126/science.1088759
Chen S, Iversen ES, Friebel T, Finkelstein D, Weber BL, Eisen A, Peterson LE, Schildkraut JM, Isaacs C, Peshkin BN, Corio C, Leondaridis L, Tomlinson G, Dutson D, Kerber R, Amos CI, Strong LC, Berry DA, Euhus DM, Parmigiani G (2006) Characterization of BRCA1 and BRCA2 mutations in a large United States sample. J Clin Oncol 24(6):863–871. https://doi.org/10.1200/JCO.2005.03.6772
Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, Evans DG, Izatt L, Eeles RA, Adlard J, Davidson R, Eccles D, Cole T, Cook J, Brewer C, Tischkowitz M, Douglas F, Hodgson S, Walker L, Porteous ME, Morrison PJ, Side LE, Kennedy MJ, Houghton C, Donaldson A, Rogers MT, Dorkins H, Miedzybrodzka Z, Gregory H, Eason J, Barwell J, McCann E, Murray A, Antoniou AC, Easton DF, Embrace (2013) Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst 105(11):812–822. https://doi.org/10.1093/jnci/djt095
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, Jervis S, van Leeuwen FE, Milne RL, Andrieu N, Goldgar DE, Terry MB, Rookus MA, Easton DF, Antoniou AC, Brca, Consortium BC, McGuffog L, Evans DG, Barrowdale D, Frost D, Adlard J, Ong KR, Izatt L, Tischkowitz M, Eeles R, Davidson R, Hodgson S, Ellis S, Nogues C, Lasset C, Stoppa-Lyonnet D, Fricker JP, Faivre L, Berthet P, Hooning MJ, van der Kolk LE, Kets CM, Adank MA, John EM, Chung WK, Andrulis IL, Southey M, Daly MB, Buys SS, Osorio A, Engel C, Kast K, Schmutzler RK, Caldes T, Jakubowska A, Simard J, Friedlander ML, McLachlan SA, Machackova E, Foretova L, Tan YY, Singer CF, Olah E, Gerdes AM, Arver B, Olsson H (2017) Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317(23):2402–2416. https://doi.org/10.1001/jama.2017.7112
Metcalfe K, Gershman S, Lynch HT, Ghadirian P, Tung N, Kim-Sing C, Olopade OI, Domchek S, McLennan J, Eisen A, Foulkes WD, Rosen B, Sun P, Narod SA (2011) Predictors of contralateral breast cancer in BRCA1 and BRCA2 mutation carriers. Br J Cancer 104(9):1384–1392. https://doi.org/10.1038/bjc.2011.120
Domchek SM, Friebel TM, Singer CF, Evans DG, Lynch HT, Isaacs C, Garber JE, Neuhausen SL, Matloff E, Eeles R, Pichert G, Van t’veer L, Tung N, Weitzel JN, Couch FJ, Rubinstein WS, Ganz PA, Daly MB, Olopade OI, Tomlinson G, Schildkraut J, Blum JL, Rebbeck TR (2010) Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA 304(9):967–975. https://doi.org/10.1001/jama.2010.1237
Rebbeck TR, Friebel T, Lynch HT, Neuhausen SL, van ‘t Veer L, Garber JE, Evans GR, Narod SA, Isaacs C, Matloff E, Daly MB, Olopade OI, Weber BL (2004) Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J Clin Oncol 22(6):1055–1062. https://doi.org/10.1200/JCO.2004.04.188
Hartmann LC, Sellers TA, Schaid DJ, Frank TS, Soderberg CL, Sitta DL, Frost MH, Grant CS, Donohue JH, Woods JE, McDonnell SK, Vockley CW, Deffenbaugh A, Couch FJ, Jenkins RB (2001) Efficacy of bilateral prophylactic mastectomy in BRCA1 and BRCA2 gene mutation carriers. J Natl Cancer Inst 93(21):1633–1637
Meijers-Heijboer H, van Geel B, van Putten WL, Henzen-Logmans SC, Seynaeve C, Menke-Pluymers MB, Bartels CC, Verhoog LC, van den Ouweland AM, Niermeijer MF, Brekelmans CT, Klijn JG (2001) Breast cancer after prophylactic bilateral mastectomy in women with a BRCA1 or BRCA2 mutation. N Engl J Med 345(3):159–164. https://doi.org/10.1056/NEJM200107193450301
Heemskerk-Gerritsen BA, Menke-Pluijmers MB, Jager A, Tilanus-Linthorst MM, Koppert LB, Obdeijn IM, van Deurzen CH, Collee JM, Seynaeve C, Hooning MJ (2013) Substantial breast cancer risk reduction and potential survival benefit after bilateral mastectomy when compared with surveillance in healthy BRCA1 and BRCA2 mutation carriers: a prospective analysis. Ann Oncol 24(8):2029–2035. https://doi.org/10.1093/annonc/mdt134
Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, Wardley A, Mitchell G, Earl H, Wickens M, Carmichael J (2010) Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376(9737):235–244. https://doi.org/10.1016/S0140-6736(10)60892-6
Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, Janizek JD, Huang X, Starita LM, Shendure J (2018) Accurate classification of BRCA1 variants with saturation genome editing. Nature 562(7726):217–222. https://doi.org/10.1038/s41586-018-0461-z
Sun J, Meng H, Yao L, Lv M, Bai J, Zhang J, Wang L, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Xie Y (2017) Germline mutations in cancer susceptibility genes in a large series of unselected breast cancer patients. Clin Cancer Res 23(20):6113–6119. https://doi.org/10.1158/1078-0432.CCR-16-3227
den Dunnen JT, Antonarakis SE (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15(1):7–12. https://doi.org/10.1002/(SICI)1098-1004(200001)15:1<7:AID-HUMU4>3.0.CO;2-N
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Freeman PJ, Hart RK, Gretton LJ, Brookes AJ, Dalgleish R (2018) VariantValidator: accurate validation, mapping, and formatting of sequence variation descriptions. Hum Mutat 39(1):61–68. https://doi.org/10.1002/humu.23348
Wang C, Zhang J, Wang Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Xie Y (2015) Prevalence of BRCA1 mutations and responses to neoadjuvant chemotherapy among BRCA1 carriers and non-carriers with triple-negative breast cancer. Ann Oncol 26(3):523–528. https://doi.org/10.1093/annonc/mdu559
Lee MS, Green R, Marsillac SM, Coquelle N, Williams RS, Yeung T, Foo D, Hau DD, Hui B, Monteiro AN, Glover JN (2010) Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res 70(12):4880–4890. https://doi.org/10.1158/0008-5472.CAN-09-4563
Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, Marceau C, Janssen H, Carette JE, Bennett KL, Colinge J, Superti-Furga G, Brummelkamp TR (2015) Gene essentiality and synthetic lethality in haploid human cells. Science 350(6264):1092–1096. https://doi.org/10.1126/science.aac7557
Steffensen AY, Dandanell M, Jonson L, Ejlertsen B, Gerdes AM, Nielsen FC, Hansen T (2014) Functional characterization of BRCA1 gene variants by mini-gene splicing assay. Eur J Hum Genet 22(12):1362–1368. https://doi.org/10.1038/ejhg.2014.40
Park JS, Nam EJ, Park HS, Han JW, Lee JY, Kim J, Kim TI, Lee ST (2017) Identification of a Novel BRCA1 Pathogenic Mutation in Korean Patients Following Reclassification of BRCA1 and BRCA2 Variants According to the ACMG Standards and Guidelines Using Relevant Ethnic Controls. Cancer Res Treat 49(4):1012–1021
Lu C, Xie M, Wendl MC, Wang J, McLellan MD, Leiserson MD, Huang KL, Wyczalkowski MA, Jayasinghe R, Banerjee T, Ning J, Tripathi P, Zhang Q, Niu B, Ye K, Schmidt HK, Fulton RS, McMichael JF, Batra P, Kandoth C, Bharadwaj M, Koboldt DC, Miller CA, Kanchi KL, Eldred JM, Larson DE, Welch JS, You M, Ozenberger BA, Govindan R, Walter MJ, Ellis MJ, Mardis ER, Graubert TA, Dipersio JF, Ley TJ, Wilson RK, Goodfellow PJ, Raphael BJ, Chen F, Johnson KJ, Parvin JD, Ding L (2015) Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun 6:10086.
Zhang H, Li L, Wang Y, Yin CC, Xie Y, Liu X, Ding H, Tian Z, Shen J, He L, Xia M, Ma X, Wu L (2017) Functional analysis of BRCT missense mutations in BRCA1-mutated Chinese Han familial breast cancer. Oncol Lett 14(5):5839–5844. https://doi.org/10.3892/ol.2017.7003
Quaresima B, Faniello MC, Baudi F, Crugliano T, Di Sanzo M, Cuda G, Costanzo F, Venuta S (2006) Missense mutations of BRCA1 gene affect the binding with p53 both in vitro and in vivo. Oncol Rep 16(4):811–815. https://doi.org/10.3892/or.16.4.811
Hayes F, Cayanan C, Barillà D, Monteiro AN (2000) Functional assay for BRCA1: mutagenesis of the COOH-terminal region reveals critical residues for transcription activation. Cancer Res 60(9):2411–2418
Acknowledgements
We thank Berry Genomics (Beijing) for kind help with the multigene panel testing in this study.
Funding
This study was funded by the National Natural Science Foundation of China (81772824) and the Science Foundation of Peking University Cancer Hospital (2017-8).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. This study was designed by Yuntao Xie. Material preparation, data collection and analysis were performed by Qiting Wan, Li Hu, Ye Xu, Tao Ouyang, Jinfeng Li, Tianfeng Wang, Zhaoqing Fan, Tie Fan, Benyao Lin. The manuscript was revised by Ye Xu and Yuntao Xie. All authors commented on previous versions of the manuscript and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was reviewed and approved by the Ethics Committee of Peking University Cancer Hospital (No. 2011KT12).
Informed consent
Written informed consent was obtained from all study participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wan, Q., Hu, L., Ouyang, T. et al. Clinical phenotypes combined with saturation genome editing identifying the pathogenicity of BRCA1 variants of uncertain significance in breast cancer. Familial Cancer 20, 85–95 (2021). https://doi.org/10.1007/s10689-020-00202-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-020-00202-4