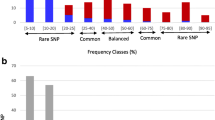
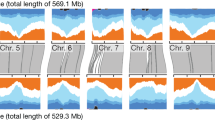
Abstract
The interior Southeastern United States could contain novel germplasm for the bioenergy crop switchgrass due to its diverse habitats and geographic location between genetic subpopulations (Atlantic, Midwest, and Gulf). Collections from this region could accelerate breeding progress, contribute to conservation efforts, and improve understanding of isolated grasslands in the region. This study located 22 sites in the Midsouth region and obtained 1,521,210 single nucleotide polymorphism markers of 202 individuals through genotype-by-sequencing. Individuals were evaluated for flowering time, winter survival and tiller number. Comparison to a national diversity panel revealed that branches of two major subpopulations occur in the region with two levels of polyploidy: Atlantic tetraploids and Midwest octoploids. Two locations contained admixed octoploid individuals with Midwest and Gulf genetics. Field performance of the Midwest octoploids conformed with prior reported performance of the Midwest subpopulation, although three sites contained promising late flowering traits. The Atlantic tetraploids had moderate winter survival, short stature, and anomalously early flowering. Atlantic populations mostly occurred in marginal sites and their morphological and flowering time adaptations may be a resource conservation strategy. Demographic inference of historical effective population size variation in a subset of tetraploid locations indicated a widespread recent decline in effective population size. This pattern is consistent with isolation of these switchgrass communities from larger populations and is further supported by evidence of inbreeding within the populations (FI = 0.18). The populations documented in this study contain novel genetic diversity and adaptations to a range of marginal habitats. Therefore, this study provides a new source of germplasm for future breeding and conservation programs.





Similar content being viewed by others
References
Alexander EB (2009) Serpentine geoecology of the eastern and southeastern margins of North America. Northeast Nat 16(sp5):223–252. https://doi.org/10.1656/045.016.0518
Ashcroft MB, Gollan JR, Warton DI, Ramp D (2012) A novel approach to quantify and locate potential microrefugia using topoclimate, climate stability, and isolation from the matrix. Glob Change Biol 18(6):1866–1879. https://doi.org/10.1111/j.1365-2486.2012.02661.x
Ayres DL, Darling A, Zwickl DJ, Beerli P, Holder MT et al (2012) BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst Biol 61(1):170–173. https://doi.org/10.1093/sysbio/syr100
Barker RE, Haas RJ, Berdahl JD, Jacobson ET (1990) Registration of ‘Dacotah’ switchgrass. Crop Sci. https://doi.org/10.2135/cropsci1990.0011183X003000050050x
Baskin JM, Baskin CC (1988) Endemism in rock outcrop plant communities of unglaciated eastern united states: an evaluation of the roles of the edaphic. Genetic and Light Factors J Biogeogr 15(5/6):829–840. https://doi.org/10.2307/2845343
Brandes E, McNunn GS, Schulte LA, Muth DJ, VanLoocke A et al (2018a) Targeted subfield switchgrass integration could improve the farm economy, water quality, and bioenergy feedstock production. GCB Bioenergy 10(3):199–212. https://doi.org/10.1111/gcbb.12481
Brandes E, Plastina A, Heaton EA (2018b) Where can switchgrass production be more profitable than corn and soybean? An integrated subfield assessment in Iowa, USA. GCB Bioenergy 10(7):473–488. https://doi.org/10.1111/gcbb.12516
Cartwright JM, Wolfe WJ (2016) Insular ecosystems of the southeastern United States—a regional synthesis to support biodiversity conservation in a changing climate. U.S. Geological Survey, Reston, VA.
Casler MD (2019) Selection for flowering time as a mechanism to increase biomass yield of upland switchgrass. BioEnergy Res. https://doi.org/10.1007/s12155-019-10044-3
Casler MD, Vogel KP (2014) Selection for biomass yield in upland, lowland, and hybrid switchgrass. Crop Sci 54(2):626. https://doi.org/10.2135/cropsci2013.04.0239
Casler MD, Vogel KP, Harrison M (2015) Switchgrass germplasm resources. Crop Sci 55(6):2463–2478. https://doi.org/10.2135/cropsci2015.02.0076
Casler MD, Vogel KP, Lee DK, Mitchell RB, Adler PR et al (2018) 30 Years of progress toward increased biomass yield of switchgrass and big bluestem. Crop Sci 58(3):1242–1254. https://doi.org/10.2135/cropsci2017.12.0729
Casler MD, Lee D, Mitchell RB, Adler PR, Sulc RM et al (2019) Nitrogen demand associated with increased biomass yield of switchgrass and big bluestem: implications for future breeding strategies. BioEnergy Res. https://doi.org/10.1007/s12155-019-10081-y
Covarrubias-Pazaran G (2016) Genome-assisted prediction of quantitative traits using the R package sommer. PLoS ONE 11(6): e0156744. https://doi.org/10.1371/journal.pone.0156744
Cullis BR, Smith AB, Coombes NE (2006) On the design of early generation variety trials with correlated data. J Agric Biol Environ Stat 11(4):381. https://doi.org/10.1198/108571106X154443
Dandine-Roulland C, Bellenguez C, Debette S, Amouyel P, Génin E et al (2016) Accuracy of heritability estimations in presence of hidden population stratification. Sci Rep 6(1):26471. https://doi.org/10.1038/srep26471
Dobrowski SZ (2011) A climatic basis for microrefugia: the influence of terrain on climate. Glob Change Biol 17(2):1022–1035. https://doi.org/10.1111/j.1365-2486.2010.02263.x
Duvick DN (1984) Genetic contributions to yield gains of U.S. hybrid maize, 1930 to 1980. Genetic contributions to yield gains of five major crop plants. Wiley, New Jersey, pp 15–47
Duvick DN (2005) Genetic progress in yield of United States maize (Zea mays L.). Maydica 50:193–202
Endelman JB (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4(3):250–255. https://doi.org/10.3835/plantgenome2011.08.0024
Estill JC, Cruzan MB (2001) Phytogeography of rare plant species endemic to the southeastern United States. Castanea 66(1/2):3–23
Evans J, Sanciangco MD, Lau KH, Crisovan E, Barry K et al (2018) Extensive genetic diversity is present within north american switchgrass germplasm. Plant Genome. https://doi.org/10.3835/plantgenome2017.06.0055
Fesenmyer KA, Christensen NL (2010) Reconstructing holocene fire history in a southern Appalachian forest using soil charcoal. Ecology 91(3):662–670. https://doi.org/10.1890/09-0230.1
Fike JH, Pease JW, Owens VN, Farris RL, Hansen JL et al (2017) Switchgrass nitrogen response and estimated production costs on diverse sites. GCB Bioenergy 9(10):1526–1542. https://doi.org/10.1111/gcbb.12444
Gelfand I, Hamilton SK, Kravchenko AN, Jackson RD, Thelen KD et al (2020) Empirical evidence for the potential climate benefits of decarbonizing light vehicle transport in the U.S. with bioenergy from purpose-grown biomass with and without BECCS. Environ Sci Technol. https://doi.org/10.1021/acs.est.9b07019
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ et al (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE 9(2):e90346. https://doi.org/10.1371/journal.pone.0090346
Godt MJW, Johnson BR, Hamrick JL (1996) Genetic diversity and population size in four rare southern Appalachian plant species. Conserv Biol 10(3):796–805
Gould BA, Palacio-Mejia JD, Jenkins J, Mamidi S, Barry K et al (2018) Population genomics and climate adaptation of a C4 perennial grass, Panicum hallii (Poaceae). BMC Genomics 19(1):792. https://doi.org/10.1186/s12864-018-5179-7
Grabowski PP, Evans J, Daum C, Deshpande S, Barry KW et al (2017) Genome-wide associations with flowering time in switchgrass using exome-capture sequencing data. New Phytol 213(1):154–169. https://doi.org/10.1111/nph.14101
Green AJ, Berger G, Griffey CA, Pitman R, Thomason W et al (2012) Genetic yield improvement in soft red winter wheat in the eastern United States from 1919 to 2009. Crop Sci 52(5):2097–2108. https://doi.org/10.2135/cropsci2012.01.0026
Hayes BJ, Bowman PJ, Chamberlain AC, Verbyla K, Goddard ME (2009) Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet Sel Evol GSE 41:51. https://doi.org/10.1186/1297-9686-41-51
Hewitt G (2000) The genetic legacy of the quaternary ice ages. Nature 405(6789):907–913. https://doi.org/10.1038/35016000
Hill JG, Hill JG, Barone JA, Barone JA, Allen C et al (2018) Southeastern grasslands: biodiversity, ecology, and management. The University of Alabama Press, Tuscaloosa
Jiang H, Lei R, Ding S-W, Zhu S (2014) Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15(1):182. https://doi.org/10.1186/1471-2105-15-182
Liu X, Fu Y-X (2020) Stairway plot 2: demographic history inference with folded SNP frequency spectra. Genome Biol 21(1):280. https://doi.org/10.1186/s13059-020-02196-9
Lovell JT, MacQueen AH, Mamidi S, Bonnette J, Jenkins J et al (2021) Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass. Nature. https://doi.org/10.1038/s41586-020-03127-1
Lowry DB, Behrman KD, Grabowski P, Morris GP, Kiniry JR et al (2014) Adaptations between ecotypes and along environmental gradients in panicum virgatum. Am Nat 183(5):682–692. https://doi.org/10.1086/675760
Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH et al (2013) Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLOS Genet 9(1):e1003215. https://doi.org/10.1371/journal.pgen.1003215
Mamidi S, Healey A, Huang P, Grimwood J, Jenkins J et al (2020) A genome resource for green millet Setaria viridis enables discovery of agronomically valuable loci. Nat Biotechnol 38(10):1203–1210. https://doi.org/10.1038/s41587-020-0681-2
McMillan C (1959) The role of ecotypic variation in the distribution of the central grassland of north America. Ecol Monogr 29(4):285–308. https://doi.org/10.2307/1942132
Morris GP, Grabowski PP, Borevitz JO (2011) Genomic diversity in switchgrass (Panicum virgatum): from the continental scale to a dune landscape. Mol Ecol 20(23):4938–4952. https://doi.org/10.1111/j.1365-294X.2011.05335.x
Nageswara-Rao M, Stewart CN, Kwit C (2013) Genetic diversity and structure of natural and agronomic switchgrass (Panicum virgatum L.) populations. Genet Resour Crop Evol 60(3):1057–1068. https://doi.org/10.1007/s10722-012-9903-x
Noss RF, Platt WJ, Sorrie BA, Weakley AS, Means DB et al (2015) How global biodiversity hotspots may go unrecognized: lessons from the north American Coastal Plain. Divers Distrib 21(2):236–244. https://doi.org/10.1111/ddi.12278
Noss RF (1995) Endangered ecosystems of the United States: a preliminary assessment of loss and degradation. USDeptof the interior, national biological service: May be obtained from the Publications Unit, USFish and Wildlife Service
Noss RF (2013) Forgotten grasslands of the south natural history and conservation. Forgotten Grasslands of the South. Island Press/Center for Resource Economics. p 1–32
Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM et al (2010) The rate and molecular spectrum of spontaneous mutations in arabidopsis thaliana. Science 327(5961):92–94. https://doi.org/10.1126/science.1180677
Peixoto MDM, Sage RF (2016) Improved experimental protocols to evaluate cold tolerance thresholds in Miscanthus and switchgrass rhizomes. GCB Bioenergy 8(2):257–268. https://doi.org/10.1111/gcbb.12301
Perrin R, Vogel K, Schmer M, Mitchell R (2008) Farm-scale production cost of switchgrass for biomass. BioEnergy Res 1(1):91–97. https://doi.org/10.1007/s12155-008-9005-y
Perrin RK, Fulginiti LE, Alhassan M (2017) Biomass from marginal cropland: willingness of North Central US farmers to produce switchgrass on their least productive fields. Biofuels Bioprod Biorefining 11(2):281–294. https://doi.org/10.1002/bbb.1741
Pitman WD (2021) Changing ecological and agricultural expectations for US coastal plain managed grasslands. Restor Ecol 29(S1):e13436. https://doi.org/10.1111/rec.13436
Poudel HP, Lee D, Casler MD (2020) Selection for Winter Survivorship in Lowland Switchgrass. BioEnergy Res 13(1):109–119. https://doi.org/10.1007/s12155-020-10091-1
Poudel, H.P., M.D. Sanciangco, S.M. Kaeppler, C.R. Buell, and M.D. Casler. (2019). Genomic Prediction for Winter Survival of Lowland Switchgrass in the Northern USA. G3amp58 GenesGenomesGenetics1:1–3.
Razar RM, Missaoui A (2018) Phenotyping winter dormancy in switchgrass to extend the growing season and improve biomass yield. J Sustain Bioenergy Syst 08:1. https://doi.org/10.4236/jsbs.2018.81001
Roley SS, Duncan DS, Liang D, Garoutte A, Jackson RD et al (2018) Associative nitrogen fixation (ANF) in switchgrass (Panicum virgatum) across a nitrogen input gradient. PLoS ONE 13(6):e0197320. https://doi.org/10.1371/journal.pone.0197320
Russell DA, Rich FJ, Schneider V, Lynch-Stieglitz J (2009) A warm thermal enclave in the Late Pleistocene of the South-eastern United States. Biol Rev 84(2):173–202. https://doi.org/10.1111/j.1469-185X.2008.00069.x
Sarath G, Baird LM, Mitchell RB (2014) Senescence, dormancy and tillering in perennial C4 grasses. Plant Sci 217–218:140–151. https://doi.org/10.1016/j.plantsci.2013.12.012
Schmidt JW (1984) Genetic contributions to yield gains in wheat. Genetic contributions to yield gains of five major crop plants. Wiley, New Jersey, pp 89–101
Schmidt P, Hartung J, Bennewitz J, Piepho H-P (2019) Heritability in plant breeding on a genotype-difference basis. Genetics 212(4):991–1008. https://doi.org/10.1534/genetics.119.302134
Schwartz C, Amasino R (2013) Nitrogen recycling and flowering time in perennial bioenergy crops. Front Plant Sci. https://doi.org/10.3389/fpls.2013.00076
Shattelroe MD, Mangum P, Ward JR, Estep MC (2021) Genetic diversity and population structure of the forgotten geum. Geum Geniculatum Michx Castanea 85(2):404–416. https://doi.org/10.2179/0008-7475.85.2.404
Tompkins RD, Stringer WC, Bridges WC (2011) An Outcrossing reciprocity study between remnant big bluestem (Andropogon gerardii) populations in the carolinas. Ecol Restor 29(4):339–345. https://doi.org/10.3368/er.29.4.339
Tompkins RD, Trapnell DW, Hamrick JL, Stringer WC (2012) Genetic variation within and among remnant big bluestem (Andropogon gerardii, Poaceae) populations in the carolinas. Southeast Nat 11(3):455–468. https://doi.org/10.1656/058.011.0309
Van Lear DH, Carroll WD, Kapeluck PR, Johnson R (2005) History and restoration of the longleaf pine-grassland ecosystem: Implications for species at risk. For Ecol Manag 211(1):150–165. https://doi.org/10.1016/j.foreco.2005.02.014
VanWallendael A, Bonnette J, Juenger TE, Fritschi FB, Fay PA et al (2020) Geographic variation in the genetic basis of resistance to leaf rust between locally adapted ecotypes of the biofuel crop switchgrass (Panicum virgatum). New Phytol 227(6):1696–1708. https://doi.org/10.1111/nph.16555
Weakley AS, Schafale MP (1994) Non-alluvial wetlands of the Southern Blue Ridge — diversity in a threatened ecosystem. Water Air Soil Pollut 77(3):359–383. https://doi.org/10.1007/BF00478428
Weigl PD, Knowles TW (2014) Temperate mountain grasslands: a climate-herbivore hypothesis for origins and persistence. Biol Rev 89(2):466–476. https://doi.org/10.1111/brv.12063
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370. https://doi.org/10.2307/2408641
Weir BS, Goudet J (2017) A unified characterization of population structure and relatedness. Genetics 206(4):2085–2103. https://doi.org/10.1534/genetics.116.198424
Zhang Y, Zalapa J, Jakubowski AR, Price DL, Acharya A et al (2011a) Natural hybrids and gene flow between upland and lowland switchgrass. Crop Sci 51(6):2626–2641. https://doi.org/10.2135/cropsci2011.02.0104
Zhang Y, Zalapa JE, Jakubowski AR, Price DL, Acharya A et al (2011b) Post-glacial evolution of panicum virgatum: centers of diversity and gene pools revealed by SSR markers and cpDNA sequences. Genetica 139(7):933. https://doi.org/10.1007/s10709-011-9597-6
Acknowledgements
NWT is grateful for the help of many who provided their time, expertise, and/or collection permits to this project: Mamie Coburn of The Nature Conservancy, Gary Kauffman of the USDA Forest Service, Kyle Pursel of Highlands-Cashiers Land Trust, David Lee with Conserving Carolinas, Wesley Sketo with the North Carolina Department of Agriculture, Rusty Boles with the Tennessee Wildlife Resource Agency, Dwayne Estes with the Southeastern Grasslands Initiative, Alex Faught of the U.S. Forest Service, Dot Fields of the Virginia Department of Conservation and Recreation, David Danley recently from the USDA Forest Service (retired), Alan Weakley, Director of the UNC Herbarium, and Robert Peet, Associate Professor at UNC. I would also like to thank Professor Aaron Ragsdale with assistance interpreting demographic inference methods.
Funding
This material is based upon work supported in part by the Great Lakes Bioenergy Research Center, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC0018409. This research was also supported by congressionally allocated funds to the USDA-ARS, U.S. Dairy Forage Research Center.
Author information
Authors and Affiliations
Contributions
NT: Conceptualization, data collection and curation, formal analysis, writing. MC: Project conception and direction, funding acquisition, supervision, methodology, review and editing of manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Data availability
Unfiltered SNP calls (in variant call format) and field performance BLUPs will be posted to Dryad Digital Repository. Germplasm from the collected populations can be obtained by contacting the authors and will be deposited into the USDA Germplasm Resources Information Network.
Code availability
Code used for this study can be obtained by contacting the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10722_2021_1282_MOESM2_ESM.png
Read count distributions of reference alleles within heterozygote calls for each individual by location. Tetraploids with disomic inheritance should display a peak at 0.50. Octoploid individuals with tetrasomic inheritance should display a peak at 0.75 (PNG 156 kb)
10722_2021_1282_MOESM3_ESM.png
A genetic distance dendrogram constructed using UPGMA (average) hierarchical clustering. Analysis included 731 individuals (202 from the Midsouth collections and 529 from the Lovell et al. diversity panel). A total of 109,729 SNPs were used to calculate the genetic distance among individuals. Individuals from the diversity panel were labeled by the state of origin, while the Midsouth collections were labeled with a site code (PNG 472 kb)
Rights and permissions
About this article
Cite this article
Tilhou, N.W., Casler, M.D. Surveying Grassland Islands: the genetics and performance of Appalachian switchgrass (Panicum virgatum L.) collections. Genet Resour Crop Evol 69, 1039–1055 (2022). https://doi.org/10.1007/s10722-021-01282-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01282-6