Abstract
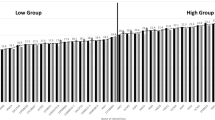
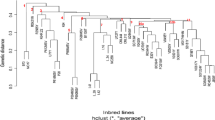
Waxy maize (Zea mays L. var. ceratina Kulesh) is an important economic crop in China, most of which are distributed in Southwestern China. In this study, 30 main waxy maize inbred lines in Southwest China were used as materials, and genetic analysis of the tested waxy corn materials was carried out using high-quality single nucleotide polymorphism (SNP) marker technology. A total of 15,111 SNPs were obtained from 30 test materials, and the genetic similarity coefficient varied from 0.4568 to 0.9974. The results of population genetic structure and principal component analysis showed that the tested waxy maize materials and the commonly used common maize classification representative inbred lines (B73, ZI330, Mo17, Huangzao 4, etc.) could not be effectively clustered. The 30 waxy corn materials can be divided into five groups separately. The application of the genetic relationship between materials identified by SNP analysis will enable breeders to select different parents to develop high-yield varieties with improved quality traits.





Similar content being viewed by others
References
Beaumont MA, Nichols RA (1996) Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond 263(1377):1619–1626
Chen ZJ (2021) Genetic diversity analysis of 87 inbred lines of waxy maize. Seed 40(6):7
Chen J, Yang YF, Guo Q (2009) Study on genetic diversity about 40 waxy corn inbred lines by SSR markers. J Maize Sci 2009
Collins GN (1909) A new type of Indian corn from China
Corrado G, Piffanelli P, Caramante M, Coppola M, Rao R (2013) SNP genotyping reveals genetic diversity between cultivated landraces and contemporary varieties of tomato. BMC Genomics 14(1):835
Evanno GS, Regnaut SJ, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Fernie AR, Tadmor Y, Zamir D (2006) Natural genetic variation for improving crop quality. Curr Opin Plant Biol 9(2):196–202
Guohua Z, Xiaowan Z, Jian M (2015) Effects of maturity on nutrients and color of chongqing sweet- waxy maize seed. J Chin Cereals Oils Assoc
Inghelandt DV, Melchinger AE, Lebreton C, Stich B (2010) Population structure and genetic diversity in a commercial maize breeding program assessed with SSR and SNP markers. Theor Appl Genet 120(7):1289–1299
Lei ZC-bYT-yWS-yZWC (2020) A new waxy corn F1 hybrid—‘Q-nuo No.5’. China Vegetables
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 9:2128–2129
Lu Y, Shah T, Hao Z, Taba S, Zhang S (2011) Comparative SNP and haplotype analysis reveals a higher genetic diversity and rapider LD decay in tropical than temperate germplasm in maize. PLoS ONE 6(9):e24861
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19(2):153–170
Rohlf F, Rohlf F, Rohlf F, Rohlf F, Rohlf FJ (2000) NTSYS-pc, Numerical Taxonomy and Multivariate Analysis System, version 2.1
Shi Y, Baishan LU, Song W, Li XU, Zhao J (2015) Genetic diversity analysis of waxy corn inbred lines by single nucleotide polymorphism (SNP) markers. Acta Agriculturae Boreali-Sinica
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Wang R, Yu Y, Zhao J, Shi Y, Song Y, Wang T, Yu L (2008) Population structure and linkage disequilibrium of a mini core set of maize inbred lines in China. Theor Appl Genet 117(7):1141–1153
Wu X, Li Y, Shi Y, Song Y, Wang T, Huang Y, Li Y (2014) Fine genetic characterization of elite maize germplasm using high-throughput SNP genotyping. Theor Appl Genet 127(3):621–631
Wu YS, Zheng YL, Sun R, Wu SY, Bi YH (2004) Genetic diversity of waxy corn and popcorn landraces in Yunnan by SSR markers. Acta Agronomica Sinica
Jin-Feng WU, Song W, Wang R, Tian HL, Xue LI, Wang FG, Zhao JR, Wei RH, Agronomy FO, University JA (2014) Heterotic grouping of 51 maize inbred lines by SNP markers. J Maize Sci 2014
Yu RH, Wang YL, Sun Y, Liu B (2012) Analysis of genetic distance by SSR in waxy maize. Genet Mol Res GMR 11(1):254–260
Zeng M (1987) Blood relationship of Chinese waxy maize
Zhang X, Zhang H, Li L, Lan H, Ren Z, Liu D, Wu L, Liu H, Jaqueth J, Li B (2016) Characterizing the population structure and genetic diversity of maize breeding germplasm in Southwest China using genome-wide SNP markers. BMC Genomics 17(1):697
Zhang SH, Yang H, Lei KR, Cai ZR, Xian H, Hong-Hua YI, Chen RL (2011) Analysis of Heterosis Groups and Heterosis models of waxy maize breeding in Chongqing. Southwest China J Agric Sci
Zheng H, Wang H, Hua Y, Wu J, Shi B, Cai R, Xu Y, Wu A, Luo L (2013) Genetic diversity and molecular evolution of Chinese waxy maize germplasm. PLoS ONE 8
Zhang Caibo YT, Wen S, Zhang W, Chen L (2021) A new waxy corn cultivar ‘Jinnuo 2’. Acta Horticulturae Sinica
Acknowledgements
This work was supported by the Chongqing Scientific Research Institution Performance Incentive Guidance Project (cstc2020jxjl80012); Chongqing Basic Scientific Research Fund Project (2016cstc-jbky-00501). The author would like to thank Ms. Chen Xiangying, Chongqing Academy of Agricultural Sciences, for her help in writing this article.
Funding
This work was supported by the Chongqing Scientific Research Institution Performance Incentive Guidance Project (cstc2020jxjl80012); and Chongqing Basic Scientific Research Fund Project (2016cstc-jbky-00501).
Author information
Authors and Affiliations
Contributions
This work was carried out in collaboration among all authors. Authors C.Z. and S.W. designed the experiments and provided resources for all experiments. Authors W.Z., L.C. and T.Y. collected the tested materials and performed field management. Author C.Z. conducted the DNA extraction. Authors C.Z., S.Z. and X.C. performed the experiments, managed the data analysis and wrote the manuscripts.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, C., Zhang, S., Chen, L. et al. Analysis of waxy maize germplasm resources in Southwest China based on SNP markers. Genet Resour Crop Evol 70, 607–616 (2023). https://doi.org/10.1007/s10722-022-01449-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01449-9