Abstract
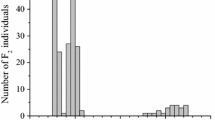
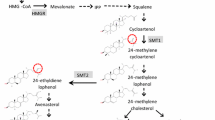
Sunflower oil with high oleic acid content is in great demand due to its nutritional as well as industrial benefits. The trait is mainly controlled by dominant alleles at a major gene, Ol, with other modifiers. The objectives of this research were to map the oil content, oleic acid and linoleic acid content in sunflower seeds. An F2 mapping population from cytoplasmic male-sterile line COSF 7A (33–35 % oleic acid) and high oleic acid inbred line HO 5–13 (88–90 % oleic acid) was developed and phenotyped for oil content, oleic acid and linoleic acid content at the F2 seed level. High phenotypic and genotypic coefficients of variation were recorded for oleic acid and linoleic acid content. High heritability and high genetic advance as percent of mean was recorded for oleic acid and linoleic acid content. This indicated the presence of the additive type of gene action controlling the traits oleic acid content and linoleic acid content. The Ol gene was mapped to linkage group (LG) 14 and tightly linked to the marker HO_Fsp_b. In addition, two more quantitative trait loci (QTLs) for oleic acid content were identified in LG8 and LG9. Two QTLs for oil content and two QTLs for linoleic acid content were also identified. All these QTLs explained over 10 % of phenotypic variation. A study was conducted with 13 genotypes differing in oil quality as well as quantity over three seasons to assess the reliability of the identified QTLs over seasons. It resulted in the identification of two potential QTLs for oleic acid as well as linoleic acid content with the markers ORS 762 and HO_Fsp_b. These markers explained more than 57.6–66.6 % of phenotypic variation. Hence it can be concluded that these markers/QTLs would be useful in the marker-assisted selection breeding programme to improve oil quality. The present study also indicated the presence of at least two other genomic regions controlling oleic and linoleic acid content in sunflower.


Similar content being viewed by others
References
Al-Chaarani RG, Gentzbittel L, Huang XQ, Sarrafi A (2004) Genotypic variation and identification of QTLs for agronomic traits, using AFLP and SSR markers in RILs of sunflower (Helianthus annuus L.). Theor Appl Genet 109(7):1353–1360
Burke JM, Knapp SJ, Rieseberg LH (2005) Genetic consequences of selection during the evolution of cultivated sunflower. Genetics 171:1933–1940
Canvin DT (1965) The effect of temperature on the oil content and fatty acid composition of the oils from several oilseed crops. Can J Bot 43:63–69
Codex Alimentarius Commission (2005) Twenty-eighth session report. FAO Headquarters, Rome, pp 4–9
Delplanque B (2000) Intéret nutritionnel des tournesols. Proceedings of XV International Sunflower Conference, Toulouse 1:15–16
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Ebrahimi A, Maury P, Berger M, Poormohammad Kiani S, Nabipour A, Shariati F, Grieu P, Sarrafi A (2008) QTL mapping of seed-quality traits in sunflower recombinant inbred lines under different water regimes. Genome 51(8):599–615
Fernandez-Martinez J, Jimenez A, Dominguez J, Garcia JM, Garces R, Mancha M (1989) Genetic analysis of the high oleic acid content in cultivated sunflower (Helianthus annuus L.). Euphytica 41:39–51
Fuller M, Diamond J, Applewhite T (1967) High oleic sunflower oil stability and chemical modification. J Am Oil Chem Soc 44:264–267
Grundy SM (1986) Comparison of monounsaturated fatty acids and carbohydrates for lowering plasma cholesterol. N Engl J Med 314:745–748
Hongtrakul V, Slabaugh MB, Knapp SJ (1998) A seed specific delta-12 oleate desaturase gene is duplicated, rearranged, and weakly expressed in high oleic acid sunflower lines. Crop Sci 38(5):1245–1249
Hu J (2006) Defining the sunflower (Helianthus annuus L.) linkage group ends with the Arabidopsis-type telomere sequence repeat-derived markers. Chromosome Res 14:535–548
Johnson HW, Robinson HF, Comstock RE (1955) Estimates of genetic and environmental variability in soybean. Agron J 47:314–318
Kinman ML, Earle FR (1964) Agronomic performance and chemical composition of the seed of sunflower hybrids and introduced varieties. Crop Sci 4:417–420
Lacombe S, Berville A (2001) A dominant mutation for high oleic acid content in sunflower (Helianthus annuus L.) seed oil is genetically linked to a single oleate-desaturase RFLP locus. Mol Breed 8(2):129–137
Lacombe S, Souyris I, Berville AJ (2009) An insertion of oleate desaturase homologous sequence silences via siRNA the functional gene leading to high oleic acid content in sunflower seed oil. Mol Genet Genom 281:43–54
Miller JF, Zimmerman DC, Vick BA (1987) Genetic control of high oleic acid content in sunflower oil. Crop Sci 27(5):923–926
Perez-Vich B, Fernandez-Martinez JM, Grondona M, Knapp SJ, Berry ST (2002) Stearoyl-ACP and oleoyl-PC desaturase genes cosegregate with quantitative trait loci underlying high stearic and high oleic acid mutant phenotypes in sunflower. Theor Appl Genet 104(2–3):338–349
Schuppert GF, Tang S, Slabaugh MB, Knapp SJ (2006) The sunflower high-oleic mutant Ol carries variable tandem repeats of FAD2-1, a seed specific oleoyl-phosphatidyl choline desaturase. Mol Breed 17:214–256
Soldatov KI (1976) Chemical mutagenesis in sunflower breeding. In: Proceedings of 7th international sunflower association, Vlaardingen, The Netherlands, pp 352–357
Voorrips RE (2006) MapChart 2.2: software for the graphical presentation of linkage maps and QTLs. Plant Research International, Wageningen
Zeng ZB (1993) Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. In: Proceedings of the National Academy of Sciences of the USA, pp 10972–10976
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136(4):1457–1468
Acknowledgement
Ameena Premnath was supported by INSPIRE fellowship (IF 120124) of Department of Science and Technology, Government of India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Premnath, A., Narayana, M., Ramakrishnan, C. et al. Mapping quantitative trait loci controlling oil content, oleic acid and linoleic acid content in sunflower (Helianthus annuus L.). Mol Breeding 36, 106 (2016). https://doi.org/10.1007/s11032-016-0527-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-016-0527-2