Abstract
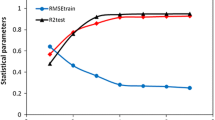
Structural analysis of topoisomerase IIα catalytic inhibitors exhibited anti-tumor properties to use them in cancer therapeutic procedures. In this study, a quantitative structure-activity relationship (QSAR) modeling including multiple linear regression (MLR) and least squares-support vector machine (LS-SVM) analysis was applied on a series of 46 synthesized xanthone derivatives as topoisomerase IIα inhibitors. It was aimed to predict half-maximal inhibitory activity (IC50) of the compounds against breast cancer cell line HCT15 using the best computational method with the least error prediction. Genetic algorithm multiple linear regression (GA-MLR) explored the functional parameters of the final models. The achieved QSAR models presented a reliable relationship between chemical structure properties as descriptors and the inhibitory activities of compounds. The models were confirmed using Leave one out-cross-validation (LOO-CV) method (for the best linear model including whole dataset R2 = 0.955, Q2 = 0.930, RMSE = 0.151, F = 115.593 and for the best non-linear model R2 = 0.981, Q2 = 0.971, RMSE = 0.099, F = 258.902). So, it was demonstrated that these models especially non-linear models are predictive models of high quality that can produce appropriate inhibitory activity prediction of the compounds to use them in pharmaceutical industries.

Graphical abstract








Similar content being viewed by others
References
Fadaka A, Ajiboye B, Ojo O, Adewale O, Olayide I, Emuowhochere R (2017) Biology of glucose metabolization in cancer cells. J Oncol Sci 3(2):45–51. https://doi.org/10.1016/j.jons.2017.06.002
McDonald LA, Eldredge GS, Barrows LR, Ireland CM Inhibition of topoisomerase II catalytic activity by pyridoacridine alkaloids from a Cystodytes sp. ascidian: a mechanism for the apparent intercalator-induced inhibition of topoisomerase II. J Med Chem 37(22):3819–3827. https://doi.org/10.1021/jm00048a017
Park S, Hong E, Kwak SY, Jun KY, Lee ES, Kwon Y, Na Y (2016) Synthesis and biological evaluation of C1-O-substituted-3-(3-butylamino-2-hydroxy-propoxy)-xanthen-9-one as topoisomerase IIα catalytic inhibitors. Eur J Med Chem 123:211–225. https://doi.org/10.1016/j.ejmech.2016.07.046
Larsen AK, Escargueil AE, Skladanowski A (2003) Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol Ther 99(2):167–181. https://doi.org/10.1016/S0163-7258(03)00058-5
Khaheshi S, Riahi S, Mohammadi-Khanaposhtani M, Shokrollahzadeh H (2019) Prediction of amines capacity for carbon dioxide absorption based on structural characteristics. Ind Eng Chem Res 58(20):8763–8771. https://doi.org/10.1021/acs.iecr.9b00567
Gorji AE, Gorji ZE, Riahi S (2017) Quantitative structure-property relationship (QSPR) for prediction of CO 2 Henry’s law constant in some physical solvents with consideration of temperature effects. Korean J Chem Eng 34(5):1405–1415. https://doi.org/10.1007/s11814-017-0018-0
Rezaei B, Riahi S (2016) Prediction of CO2 loading of amines in carbon capture process using membrane contactors: a molecular modeling. J Nat Gas Sci Eng 33:388–396. https://doi.org/10.1016/j.jngse.2016.05.003
Mavaddat M, Riahi S (2016) A molecular structure based model for predicting optimal salinity of anionic surfactants. Fluid Phase Equilib 409:354–360. https://doi.org/10.1016/j.fluid.2015.10.010
Momeni M, Riahi S (2015) An investigation into the relationship between molecular structure and rich/lean loading of linear amine-based CO2 absorbents. Int J Greenh Gas Control 42:157–164. https://doi.org/10.1016/j.ijggc.2015.07.037
Momeni M, Riahi S (2014) Prediction of amines capacity for carbon dioxide absorption in gas sweetening processes. J Nat Gas Sci Eng 21:442–450. https://doi.org/10.1016/j.jngse.2014.09.002
He G, Feng L, Chen H (2012) A QSAR study of the acute toxicity of halogenated phenols. Procedia Eng 43:204–209. https://doi.org/10.1016/j.proeng.2012.08.035
Rezaei B, Riahi S, Gorji AE (2020) Molecular investigation of amine performance in the carbon capture process: least squares support vector machine approach. Korean J Chem Eng 37(1):72–79. https://doi.org/10.1007/s11814-019-0408-6
Mehraein I, Riahi S (2017) The QSPR models to predict the solubility of CO2 in ionic liquids based on least-squares support vector machines and genetic algorithm-multi linear regression. J Mol Liq 225:521–530. https://doi.org/10.1016/j.molliq.2016.10.133
Abbasi-Radmoghaddam Z, Riahi S, Gharaghani S, Mohammadi-Khanaposhtanai M (2020) Design of potential anti-tumor PARP-1 inhibitors by QSAR and molecular modeling studies. Mol Divers:1–15. https://doi.org/10.1007/s11030-020-10063-9
Bak A, Kozik V, Walczak M, Fraczyk J, Kaminski Z, Kolesinska B, Smolinski A, Jampilek J (2018) Towards intelligent drug design system: application of artificial dipeptide receptor library in QSAR-oriented studies. Molecules 23(8):1964. https://doi.org/10.3390/molecules23081964
Maicheen C, Phosrithong N, Jittikoon J, Ungwitayatorn J (2018) Topoisomerase I inhibitory activity and 3D QSAR studies of chromone derivatives. Chiang Mai J Science 45(2):1073–1086
Luo Y, Zhou Y, Song Y, Chen G, Wang YX, Tian Y, Fan WW, Yang YS, Cheng T, Zhu HL (2018) Optimization of substituted cinnamic acyl sulfonamide derivatives as tubulin polymerization inhibitors with anticancer activity. Bioorg Med Chem Lett 28(23–24):3634–3638. https://doi.org/10.1016/j.bmcl.2018.10.037
Masand VH, El-Sayed NN, Bambole MU, Quazi SA (2018) Multiple QSAR models, pharmacophore pattern and molecular docking analysis for anticancer activity of α, β-unsaturated carbonyl-based compounds, oxime and oxime ether analogues. J Mol Struct 1157:89–96. https://doi.org/10.1016/j.molstruc.2017.12.045
Yuanita EM, Pranowo HD, Jumina JU, Mustofa MU (2016) Design of hydroxy xanthones derivatives as anticancer using quantitative structure-activity relationship. Asian J Pharm Clin Res 9(2):3–8
Abdelhaleem EF, Abdelhameid MK, Kassab AE, Kandeel MM (2018) Design and synthesis of thienopyrimidine urea derivatives with potential cytotoxic and pro-apoptotic activity against breast cancer cell line MCF-7. Eur J Med Chem 143:1807–1825. https://doi.org/10.1016/j.ejmech.2017.10.075
Karki R, Jun KY, Kadayat TM, Shin S, Magar TB, Bist G, Shrestha A, Na Y, Kwon Y, Lee ES (2016) A new series of 2-phenol-4-aryl-6-chlorophenyl pyridine derivatives as dual topoisomerase I/II inhibitors: synthesis, biological evaluation and 3D-QSAR study. Eur J Med Chem 113:228–245. https://doi.org/10.1016/j.ejmech.2016.02.050
Release H. (2002) 7.5 for Windows. Molecular modeling system, Hypercube Inc http://www hyper com
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H. (2009). Gaussian 09; Gaussian, Inc. Wallingford, 32, 5648–5652
Todeschini R, Consonni V, Mauri A, Pavan M. (2002). DRAGON-software for the calculation of molecular descriptors version 2.1
Kerwin SM (2010). ChemBioOffice ultra 2010 suite. https://doi.org/10.1021/ja1005306
Golbraikh A, Tropsha A (2000) Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. Mol Divers 5(4):231–243. https://doi.org/10.1023/A:1021372108686
Mercader AG, Duchowicz PR (2015) Enhanced replacement method integration with genetic algorithms populations in QSAR and QSPR theories. Chemom Intell Lab Syst 149:117–122. https://doi.org/10.1016/j.chemolab.2015.10.007
Anter AM, Moemen YS, Darwish A, Hassanien AE (2020) Multi-target QSAR modelling of chemo-genomic data analysis based on extreme learning machine. Knowl-Based Syst 188:104977. https://doi.org/10.1016/j.knosys.2019.104977
Ghamali M, Chtita S, Hmamouchi R, Adad A, Bouachrine M, Lakhlifi T (2016) The inhibitory activity of aldose reductase of flavonoid compounds: combining DFT and QSAR calculations. J Taibah Univ Sci 10(4):534–542. https://doi.org/10.1016/j.jtusci.2015.09.006
Sharifi M, Ghadamyari M, Gholivand K, Valmoozi AA, Sajedi RH (2017) Characterization of acetylcholinesterase from elm left beetle, Xanthogaleruca luteola and QSAR of temephos derivatives against its activity. Pestic Biochem Physiol 136:12–22. https://doi.org/10.1016/j.pestbp.2016.08.010
Netzeva TI, Worth AP, Aldenberg T, Benigni R, Cronin MT, Gramatica P, Jaworska JS, Kahn S, Klopman G, Marchant CA, Myatt G (2005) Current status of methods for defining the applicability domain of (quantitative) structure-activity relationships: the report and recommendations of ecvam workshop 52. Altern Lab Anim 33(2):155–173. https://doi.org/10.1177/026119290503300209
Kupcewicz B, Balcerowska-Czerniak G, Małecka M, Paneth P, Krajewska U, Rozalski M (2013) Structure–cytotoxic activity relationship of 3-arylideneflavanone and chromanone (E, Z isomers) and 3-arylflavones. Bioorg Med Chem Lett 23(14):4102–4106. https://doi.org/10.1016/j.bmcl.2013.05.044
Katritzky AR, Pacureanu LM, Slavov SH, Dobchev DA, Karelson M (2008) QSPR study of critical micelle concentrations of nonionic surfactants. Ind Eng Chem Res 47(23):9687–9695. https://doi.org/10.1021/ie800954k
Ghaslani D, Gorji ZE, Gorji AE, Riahi S (2017) Chemical engineering research and design descriptive and predictive models for Henry ’ s law constant of CO 2 in ionic liquids : a QSPR study. Chem Eng Res Des 120:15–25. https://doi.org/10.1016/j.cherd.2016.12.020
Yap CW, Chen YZ (2005) Quantitative structure-pharmacokinetic relationships for drug distribution properties by using general regression neural network. J Pharm Sci 94(1):153–168. https://doi.org/10.1002/jps.20232
Ma S, Lv M, Zhang X, Zhai H, Lv W (2015) Computational study of the effects of cations and anions to the cytotoxicity of diverse ionic liquids by supervised machine learning. Chemom Intell Lab Syst 144:138–147. https://doi.org/10.1016/j.chemolab.2015.03.014
Daghir-Wojtkowiak E, Wiczling P, Bocian S, Kubik Ł, Kośliński P, Buszewski B, Kaliszan R, Markuszewski MJ (2015) Least absolute shrinkage and selection operator and dimensionality reduction techniques in quantitative structure retention relationship modeling of retention in hydrophilic interaction liquid chromatography. J Chromatogr A 1403:54–62. https://doi.org/10.1016/j.chroma.2015.05.025
Zheng F, Zheng G, Deaciuc AG, Zhan CG, Dwoskin LP, Crooks PA (2007) Computational neural network analysis of the affinity of lobeline and tetrabenazine analogs for the vesicular monoamine transporter-2. Bioorg Med Chem 15(8):2975–2992. https://doi.org/10.1016/j.bmc.2007.02.013
Zhang J, Nan X, Yu HT, Cheng PL, Zhang Y, Liu YQ, Zhang SY, Hu GF, Liu H, Chen AL (2016) Synthesis, biological activities and structure− activity relationships for new avermectin analogues. Eur J Med Chem 121:422–432. https://doi.org/10.1016/j.ejmech.2016.05.056
Kupcewicz B, Małecka M, Zapadka M, Krajewska U, Rozalski M, Budzisz E (2016) Quantitative relationships between structure and cytotoxic activity of flavonoid derivatives. An application of Hirshfeld surface derived descriptors. Bioorg Med Chem Lett 26(14):3336–3341. https://doi.org/10.1016/j.bmcl.2016.05.038
Fahmy T (2013) XLSTAT, Version 2013. Addinsoft, Paris
Acknowledgments
The authors like to acknowledge the Institute of Petroleum Engineering (IPE), the University of Tehran for their support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rahmani, N., Abbasi-Radmoghaddam, Z., Riahi, S. et al. Predictive QSAR models for the anti-cancer activity of topoisomerase IIα catalytic inhibitors against breast cancer cell line HCT15: GA-MLR and LS-SVM modeling. Struct Chem 31, 2129–2145 (2020). https://doi.org/10.1007/s11224-020-01543-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-020-01543-7