Abstract
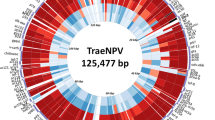
The Bombyx mori nucleopolyhedrovirus (BmNPV) La is a variant BmNPV strain isolated in Laos. La has different features from BmNPV type strain T3 in virulence, production of the polyhedrin protein, and the formation of multicapsid occlusion-derived viruses. Here, the whole-genome sequence of La was compared to the sequences of nine BmNPV and two Bombyx mandarina nucleopolyhedrovirus strains. The complete La genome consisted of 127,618 base pairs with a G + C content of 40.3% and contained putative 136 open reading frames encoding more than 60 amino acids. The La genome lacked the bro-b gene and had the highest identity with that of the T3 strain. A comparison of the transcriptomes of La- and T3-infected cells showed that the expression levels of the polyhedrin and cathepsin genes were greater in cells infected with La as compared to those infected with T3. Interestingly, the virus genes with different RNA levels between the two BmNPV strains were assembled into five clusters in the genome of La. Also, the RNA levels of host ribosomal protein genes were significantly decreased in cells infected with La as compared to those infected with T3.




Similar content being viewed by others
References
Jehle JA, Blissard GW, Bonning BC, Cory JS, Herniou EA, Rohrmann GF, Theilmann DA, Vlak JM (2006) On the classification and nomenclature of baculoviruses: a proposal for revision. Arch Virol 151:1257–1266. https://doi.org/10.1007/s00705-006-0763-6
Slack J, Arif BM (2006) The baculoviruses occlusion-derived virus: virion structure and function. Adv Virus Res 69:99–165. https://doi.org/10.1016/S0065-3527(06)69003-9
Rohrmann GF (2013) Baculovirus molecular biology; National Center for Biotechnology Information: Bethesda (MD), USA. https://www.ncbi.nlm.nih.gov/books/NBK114593/. Accessed 29 July 2019
Smith GE, Summers MD, Fraser MJ (1983) Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol 3:2156–2165. https://doi.org/10.1128/mcb.3.12.2156
Maeda S, Kawai T, Obinata M, Chika T, Horiuchi T, Maekawa K, Nakasuji K, Saeki Y, Sato Y, Yamada K, Furusawa M (1984) Characteristics of human interferon-α produced by a gene transferred by a baculovirus vector in the silkworm, Bombyx mori. Proc Jpn Acad B 60:423–426. https://doi.org/10.2183/pjab.60.423
Kadono-Okuda K, Yamamoto M, Higashino Y, Taniai K, Kato Y, Chowdhury S, Xu J, Choi SK, Sugiyama M, Nakashima K, Maeda S, Yamakawa M (1995) Baculovirus-mediated production of the human growth hormone in larvae of the silkworm, Bombyx mori. Biochem Biophys Res Commun 213:389–396. https://doi.org/10.1006/bbrc.1995.2144
Kato T, Kajikawa M, Maenaka K, Park EY (2010) Silkworm expression system as a platform technology in life science. Appl Microbiol Biotechnol 85:459–470. https://doi.org/10.1007/s00253-009-2267-2
Iwashita Y (1993) On the multiplication and antigen in variants of the NPV of silkworm, Bombyx mori. Report of the grant-in-aid for scientific research (No. 02454055) by Ministry of education, science, sports and culture (in Japanese).
Fujimoto S, Kokusho R, Kakemizu H, Izaku T, Katsuma S, Iwashita Y, Kawasaki H, Iwanaga M (2017) Characterization of a Bombyx mori nucleopolyhedrovirus variant isolated in Laos. J Insect Biotechnol Sericol 86:85–094. https://doi.org/10.11416/jibs.86.3_085
Iwanaga M, Kurihara M, Kobayashi M, Kang W (2002) Characterization of Bombyx mori nucleopolyhedrovirus orf68 gene that encodes a novel structural protein of budded virus. Virology 297:39–47. https://doi.org/10.1006/viro.2002.1443
Maeda S, Kawai T, Obinata M, Fujiwara H, Horiuchi T, Saeki Y, Furusawa M (1985) Production of human interferon in silkworm using a baculovirus vector. Nature 315:592–594. https://doi.org/10.1038/315592a0
Iwanaga M, Takaya K, Katsuma S, Ote M, Tanaka S, Kamita SG, Kang W, Shimada T, Kobayashi M (2004) Expression profiling of baculovirus genes in permissive and nonpermissive cell lines. Biochem Biophys Res Commun 323:599–614. https://doi.org/10.1016/j.bbrc.2004.08.114
Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, Ishihara G, Kawaoka S, Sugano S, Shimada T, Suzuki Y, Suzuki MG, Katsuma S (2014) A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509:633. https://doi.org/10.1038/nature13315
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Gomi S, Majima K, Maeda S (1999) Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol 80:1323–1337. https://doi.org/10.1099/0022-1317-80-5-1323
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Li D, Guo Y, Shao H, Tellier LC, Wang J, Xiang Z, Xia Q (2010) Genetic diversity, molecular phylogeny and selection evidence of the silkworm mitochondria implicated by complete resequencing of 41 genomes. BMC Evolut Biol 10:81. https://doi.org/10.1186/1471-2148-10-81
Kawamoto M, Jouraku A, Toyoda A, Yokoi K, Minakuchi Y, Katsuma S, Fujiyama A, Kiuchi T, Yamamoto K, Shimada T (2019) High-quality genome assembly of the silkworm, Bombyx mori. Insect Biochem Mol Biol 107:53–62. https://doi.org/10.1016/j.ibmb.2019.02.002
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Innami K, Aizawa T, Tsukui T, Katsuma S, Imanishi S, Kawasaki H, Iwanaga M (2016) Infection studies of nontarget mammalian cell lines with Bombyx mori macula- like virus. J Virol Methods 229:24–26. https://doi.org/10.1016/j.jviromet.2015.12.002
Daimon T, Katsuma S, Shimada T (2007) Mutational analysis of active site residues of chitinase from Bombyx mori nucleopolyhedrovirus. Virus Res 124:168–175. https://doi.org/10.1016/j.virusres.2006.11.001
Iwanaga M, Adachi Y, Uchiyama K, Tsukui K, Katsuma S, Kawasaki H (2014) Long-term adaptation of the Bombyx mori BmN4 cell line to grow in serum-free culture. Vitro Cell Dev Biol Anim 50:792–796. https://doi.org/10.1007/s11626-014-9781-y
Garavaglia MJ, Miele SA, Iserte JA, Belaich MN, Ghiringhelli PD (2012) The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae. J Virol 86:12069–12079. https://doi.org/10.1128/JVI.01873-12
Javed MA, Biswas S, Willis LG, Harris S, Pritchard C, van Oers MM, Donly BC, Erlandson MA, Hegedus DD, Theilmann DA (2017) Autographa californica multiple nucleopolyhedrovirus AC83 is a per os infectivity factor (PIF) protein required for occlusion-derived virus (ODV) and budded virus nucleocapsid assembly as well as assembly of the PIF complex in ODV envelopes. J Virol 91:e02115–e2116. https://doi.org/10.1128/JVI.02115-16
Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD (1994) The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586–605. https://doi.org/10.1006/viro.1994.1380
Kuzio J, Pearson MN, Harwood SH, Funk CJ, Evans JT, Slavicek JM, Rohrmann GF (1999) Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology 253:17–34. https://doi.org/10.1006/viro.1998.9469
Guarino LA, Summers MD (1986) Interspersed homologous DNA of Autographa californica nuclear polyhedrosis virus enhances delayed-early gene expression. J Virol 60:215–223
Bossert M, Carstens EB (2018) Sequential deletion of AcMNPV homologous regions leads to reductions in budded virus production and late protein expression. Virus Res 256:125–133. https://doi.org/10.1016/j.virusres.2018.08.010
Ohkawa T, Majima K, Maeda S (1994) A cysteine protease encoded by the baculovirus Bombyx mori nuclear polyhedrosis virus. J Virol 68:6619–6625
Hawtin RE, Zarkowska T, Arnold K, Thomas CJ, Gooday GW, King LA, Kuzio JA, Possee RD (1997) Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 238:243–253. https://doi.org/10.1006/viro.1997.8816
Pike CE (1970) Activities by country, Laos. In: Japanese efforts to diversify sources of agricultural imports. U S Department of Agriculture Economic Research Service, Washington, pp 17
Ardisson-Araújo DM, Melo FL, de Souza AM, Brancalhão RM, Báo SN, Ribeiro BM (2014) Complete genome sequence of the first non-Asian isolate of Bombyx mori nucleopolyhedrovirus. Virus Genes 49:477–484. https://doi.org/10.1007/s11262-014-1112-6
Xu YP, Cheng RL, Xi Y, Zhang CX (2013) Genomic diversity of Bombyx mori nucleopolyhedrovirus strains. Genomics 102:63–71. https://doi.org/10.1016/j.ygeno.2013.04.015
Zemskov EA, Kang W, Maeda S (2000) Evidence for nucleic acid binding ability and nucleosome association of Bombyx mori nucleopolyhedrovirus BRO proteins. J Virol 74:6784–6789. https://doi.org/10.1128/jvi.74.15.6784-6789.2000
Kotani E, Muto S, Ijiri H, Mori H (2015) Bombyx mori nucleopolyhedrovirus nucleic acid binding proteins BRO-B and BRO-E associate with host T-cell intracellular antigen 1 homologue BmTRN-1 to influence protein synthesis during infection. J Gen Virol 96:1947–1956. https://doi.org/10.1099/vir.0.000136
Moldován N, Tombácz D, Szűcs A, Csabai Z, Balázs Z, Kis E, Molnár J, Boldogkői Z (2018) Third-generation sequencing reveals extensive polycistronism and transcriptional overlapping in a baculovirus. Sci Rep 8:8604. https://doi.org/10.1038/s41598-018-26955-8
Lu A, Miller LK (1995) The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol 69:975–982
Rapp JC, Wilson JA, Miller LK (1998) Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J Virol 72:10197–10206
Lu A, Carstens EB (1993) Immediate-early baculovirus genes transactivate the p143 gene promoter of Autographa californica nuclear polyhedrosis virus. Virology 195:710–718. https://doi.org/10.1006/viro.1993.1422
Yang S, Miller LK (1999) Activation of baculovirus very late promoters by interaction with very late factor 1. J Virol 73:3404–3409
Nie Y, Fang M, Theilmann DA (2009) AcMNPV AC16 (DA26, BV/ODV-E26) regulates the levels of IE0 and IE1 and binds to both proteins via a domain located within the acidic transcriptional activation domain. Virology 385:484–495. https://doi.org/10.1016/j.virol.2008.12.020
Oppenheimer DI, Volkmann LE (1997) Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch Virol 142:2107–2113. https://doi.org/10.1007/s007050050229
Volkman LE (2015) Baculoviruses and nucleosome management. Virology 476:257–263. https://doi.org/10.1016/j.virol.2014.12.022
Mikhailov VS, Rohrmann GF (2002) Binding of the baculovirus very late expression factor 1 (VLF-1) to different DNA structures. BMC Mol Biol 3:14. https://doi.org/10.1186/1471-2199-3-14
Katsuma S, Noguchi Y, Zhou CL, Kobayashi M, Maeda S (1999) Characterization of the 25K FP gene of the baculovirus Bombyx mori nucleopolyhedrovirus: implications for post-mortem host degradation. J Gen Virol 80:783–791. https://doi.org/10.1099/0022-1317-80-3-783
Katsuma S, Nakanishi T, Shimada T (2009) Bombyx mori nucleopolyhedrovirus FP25K is essential for maintaining a steady-state level of v-cath expression throughout the infection. Virus Res 140:155–160. https://doi.org/10.1016/j.virusres.2008.11.014
Nakanishi T, Goto C, Kobayashi M, Kang W, Suzuki T, Dohmae N, Matsumoto S, Shimada T, Katsuma S (2010) Comparative studies of lepidopteran baculovirus-specific protein FP25K: development of a novel Bombyx mori nucleopolyhedrovirus-based vector with a modified fp25K gene. J Virol 84:5191–5200. https://doi.org/10.1128/JVI.00099-10
Liu X, Chen K, Cai K, Yao Q (2008) Determination of protein composition and host-derived proteins of Bombyx mori nucleopolyhedrovirus by 2-dimensional electrophoresis and mass spectrometry. Intervirology 51:369–376. https://doi.org/10.1159/000193462
Xue J, Qiao N, Zhang W, Cheng RL, Zhang XQ, Bao YY, Xu YP, Gu LZ, Han JDJ, Zhang CX (2012) Dynamic interactions between Bombyx mori nucleopolyhedrovirus and its host cells revealed by transcriptome analysis. J Virol 86:7345–7359. https://doi.org/10.1128/JVI.07217-12
Chen YW, Wu CP, Wu TC, Wu YL (2018) Analyses of the transcriptome of Bombyx mori cells infected with either BmNPV or AcMNPV. J Asia-Pacific Entomol 21:37–45. https://doi.org/10.1016/j.aspen.2017.10.009
Cheng RL, Xu YP, Zhang CX (2012) Genome sequence of a Bombyx mori nucleopolyhedrovirus strain with cubic occlusion bodies. J Virol 86:10245. https://doi.org/10.1128/JVI.01639-12
Fan HW, Zhang XC, Xu YP, Cheng XW, Zhang CX (2012) Genome of a Bombyx mori nucleopolyhedrovirus strain isolated from India. J Virol 86:11941. https://doi.org/10.1128/JVI.02040-12
Xu YP, Ye ZP, Niu CY, Bao YY, Wassng WB, Shen WD, Zhang CX (2010) Comparative analysis of the genomes of Bombyx mandarina and Bombyx mori nucleopolyhedroviruses. J Microbiol 48:102–110. https://doi.org/10.1007/s12275-009-0197-4
Xu YP, Gu LZ, Lou YH, Cheng RL, Xu HJ, Wang WB, Zhang CX (2012) A baculovirus isolated from wild silkworm encompasses the host ranges of Bombyx mori nucleopolyhedrosis virus and Autographa californica multiple nucleopolyhedrovirus in cultured cells. J Gen Virol 93:2480–2489. https://doi.org/10.1099/vir.0.043836-0
Acknowledgements
This work was supported by Grants from JSPS KAKENHI (Grant No. 19J13817) and Young Innovation Scholarship of Utsunomiya University to SF, MEXT KAKENHI (Grant No. 221S0002) to YS, JSPS KAKENHI (Grant Nos. 25292196, 16H05051, 19H02966) to SK, and Venture Business Laboratory Program F of Utsunomiya University and A-step Feasibility Study Program of the Japan Science and Technology Agency (Grant No. AS262Z01406N) to MI. The authors would like to thank Enago (www.enago.jp) for the English language review.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. The experiments were conceived and designed by Shota Fujimoto and Masashi Iwanaga. The experiments were performed by Shota Fujimoto, Munetaka Kawamoto, Keisuke Shoji, Yutaka Suzuki, Susumu Katsuma, and Masashi Iwanaga. The paper was written by Shota Fujimoto, Susumu Katsuma, and Masashi Iwanaga.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was performed in accordance with institutional committee protocols of Utsunomiya University.
Research involving human and animal participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Edited by A. Lorena Passarelli.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11262_2019_1727_MOESM1_ESM.pptx
Supplementary file1 (PPTX 844 kb)—Fig. S1 Validation of differential virus gene expression levels between La and T3. BmN cells were inoculated with BmNPV La or T3 and total RNA was purified at 72 and 96 hpi. The RNA levels of representative virus genes contained within clusters A to E were analyzed by RT-qPCR. The results are presented as the average of three independent experiments. The error bars indicate standard deviations. The asterisks indicate significant differences (Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.005). Fig. S2 Validation of MA plot analysis between La- and T3-infected cells. (a) Genes that were abundantly transcribed (A > 6) with lower RNA levels in cells infected with La as compared to those infected with T3. (b) Genes that were abundantly transcribed (A > 6) with the same RNA levels in La- and T3-infected cells. BmN cells infected with La or T3 were collected at 72 and 96 hpi, and total RNA was purified and subjected to RT-qPCR analysis. The results are presented as the average of three independent experiments. The error bars indicate standard deviations. The asterisks indicate significant differences (Student’s t-test, *p < 0.05, ***p < 0.005)
Rights and permissions
About this article
Cite this article
Fujimoto, S., Kawamoto, M., Shoji, K. et al. Whole-genome sequencing and comparative transcriptome analysis of Bombyx mori nucleopolyhedrovirus La strain. Virus Genes 56, 249–259 (2020). https://doi.org/10.1007/s11262-019-01727-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-019-01727-2