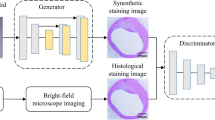
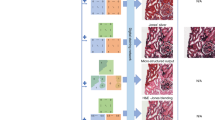
Abstract
Purpose
Histological analysis of human carotid atherosclerotic plaques is critical in understanding atherosclerosis biology and developing effective plaque prevention and treatment for ischemic stroke. However, the histological staining process is laborious, tedious, variable, and destructive to the highly valuable atheroma tissue obtained from patients.
Procedures
We proposed a deep learning-based method to simultaneously transfer bright-field microscopic images of unlabeled tissue sections into equivalent multiple sections of the same samples that are virtually stained. Using a pix2pix model, we trained a generative adversarial neural network to achieve image-to-images translation of multiple stains, including hematoxylin and eosin (H&E), picrosirius red (PSR), and Verhoeff van Gieson (EVG) stains.
Results
The quantification of evaluation metrics indicated that the proposed approach achieved the best performance in comparison with other state-of-the-art methods. Further blind evaluation by board-certified pathologists demonstrated that the multiple virtual stains have high consistency with standard histological stains. The proposed approach also indicated that the generated histopathological features of atherosclerotic plaques, such as the necrotic core, neovascularization, cholesterol crystals, collagen, and elastic fibers, are optimally matched with those of standard histological stains.
Conclusions
The proposed approach allows for the virtual staining of unlabeled human carotid plaque tissue images with multiple types of stains. In addition, it identifies the histopathological features of atherosclerotic plaques in the same tissue sample, which could facilitate the development of personalized prevention and other interventional treatments for carotid atherosclerosis.






Similar content being viewed by others
Code Availability
References
WHO The top 10 causes of death, Fact Sheet. https://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed 9 December 2020).
Pelisek J, Well G, Reeps C et al (2012) Neovascularization and angiogenic factors in advanced human carotid artery stenosis. Circ J 76:1274–1282
Pelisek J, Pongratz J, Deutsch L, Reeps C, Stadlbauer T, Eckstein HH (2012) Expression and cellular localization of metalloproteases ADAMs in high graded carotid artery lesions. Scand J Clin Lab Inv 72:648–656
Zhong XY, Ma ZC, Su YS et al (2020) Flavin adenine dinucleotide ameliorates hypertensive vascular remodeling via activating short chain acyl-CoA dehydrogenase. Life Sci 258:118156
Rivenson Y, de Haan K, Wallace WD, Ozcan A (2020) Emerging advances to transform histopathology using virtual staining. BME Frontiers 2020:1–11
Croce AC, Bottiroli G (2014) Autofluorescence spectroscopy and imaging: a tool for biomedical research and diagnosis. Eur J Histochem 58:2461
Jamme F, Kascakova S, Villette S et al (2013) Deep UV autofluorescence microscopy for cell biology and tissue histology. Biol Cell 105:277–288
Le TT, Langohr IM, Locker MJ, Sturek M, Cheng JX (2007) Label-free molecular imaging of atherosclerotic lesions using multimodal nonlinear optical microscopy. J Biomed Opt 12:54007
Zoumi A, Yeh A, Tromberg BJ (2002) Imaging cells and extracellular matrix In vivo by using second-harmonic generation and two-photon excited fluorescence. P Natl Acad Sci USA 99:11014–11019
Witte S, Negrean A, Lodder JC et al (2011) Label-free live brain imaging and targeted patching with third-harmonic generation microscopy. Proc Natl Acad Sci U S A 108:5970–5975
Ji M, Orringer DA, Freudiger CW et al (2013) Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci Transl Med 5:201ra119
Orringer DA, Pandian B, Niknafs YS et al (2017) Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng 1:27
Seeger M, Karlas A, Soliman D, Pelisek J, Ntziachristos V (2016) Multimodal optoacoustic and multiphoton microscopy of human carotid atheroma. Photoacoustics 4:102–111
Bayramoglu N, Kaakinen M, Eklund L, Heikkila J (2017) Towards virtual H&E staining of hyperspectral lung histology images using conditional generative adversarial networks. Ieee Int Conf Comp V:64–71.
Rivenson Y, Wang H, Wei Z et al (2019) Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat Biomed Eng 3:466–477
Rivenson Y, Liu TR, Wei ZS, Zhang Y, de Haan K, Ozcan A (2019) PhaseStain: the digital staining of label-free quantitative phase microscopy images using deep learning. Light-Sci Appl 8:23
Christiansen EM, Yang SJ, Ando DM et al (2018) In silico labeling: predicting fluorescent labels in unlabeled images. Cell 173:792
Liu Y, Yuan H, Wang ZY, Ji SW (2020) Global pixel transformers for virtual staining of microscopy images. Ieee T Med Imaging 39:2256–2266
Li D, Hui H, Zhang YQ et al (2020) Deep learning for virtual histological staining of bright-field microscopic images of unlabeled carotid artery tissue. Mol Imaging Biol 22:1301–1309
Zhang Y, de Haan K, Rivenson Y, Li J, Delis A, Ozcan A (2020) Digital synthesis of histological stains using micro-structured and multiplexed virtual staining of label-free tissue. Light Sci Appl 9:78
Zhou NY, Cai D, Han X, Yao JH (2019) Enhanced cycle-consistent generative adversarial network for color normalization of H&E stained images. Lect Notes Comput Sc 11764:694–702
Gupta L, Klinkhammer BM, Boor P, Merhof D, Gadermayr M (2019) GAN-based image enrichment in digital pathology boosts segmentation accuracy. Lect Notes Comput Sc 11764:631–639
Isola P, Zhu JY, Zhou TH, Efros AA (2017) Image-to-image translation with conditional adversarial networks. Proc Cvpr Ieee:5967–5976.
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. Med Imag Comput Comput Assist Interv Pt Iii 9351:234–241
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004) Image quality assessment: from error visibility to structural similarity. Ieee T Image Process 13:600–612
Zhang R, Isola P, Efros AA, Shechtman E, Wang O (2018) The unreasonable effectiveness of deep features as a perceptual metric. 2018 Ieee/Cvf Conference on Computer Vision and Pattern Recognition (Cvpr):586–595.
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun Acm 60:84–90
Choi Y, Choi M, Kim M, Ha JW, Kim S, Choo J (2018) StarGAN: unified generative adversarial networks for multi-domain image-to-image translation. 2018 Ieee/Cvf Conference on Computer Vision and Pattern Recognition (Cvpr):8789–8797.
He ZL, Zuo WM, Kan MN, Shan SG, Chen XL (2019) AttGAN: facial attribute editing by only changing what you want. Ieee T Image Process 28:5464–5478
Liu M, Ding YK, Xia M, et al. (2019) STGAN: a unified selective transfer network for arbitrary image attribute editing. 2019 Ieee/Cvf Conference on Computer Vision and Pattern Recognition (Cvpr 2019):3668–3677.
Wang W, Zhang Y, Hui H et al (2021) The effect of endothelial progenitor cell transplantation on neointimal hyperplasia and reendothelialisation after balloon catheter injury in rat carotid arteries. Stem Cell Res Ther 12:99
Tong W, Hui H, Shang W et al (2021) Highly sensitive magnetic particle imaging of vulnerable atherosclerotic plaque with active myeloperoxidase-targeted nanoparticles. Theranostics 11:506–521
Acknowledgements
The authors would like to acknowledge the instrumental and technical support of multimodal biomedical imaging experimental platform, Institute of Automation, Chinese Academy of Sciences.
Funding
This work was supported in part by the National Key Research and Development Program of China under Grant 2017YFA0700401, 2016YFC0103803; the National Natural Science Foundation of China under Grant 81730050, 81827808, 62027901, 81671851, 81527805; CAS Youth Innovation Promotion Association under Grant 2018167; CAS Scientific Instrument R&D Program under Grant YJKYYQ20170075; CAS Key Technology Talent Program; and the Project of High-Level Talents Team Introduction in Zhuhai City (Zhuhai HLHPTP201703).
Author information
Authors and Affiliations
Contributions
Guanghao Zhang: Conceptualization, Methodology, Software, Writing Original draft.
Bin Ning: Conceptualization, Data acquisition, Writing—original draft.
Hui Hui: Conceptualization, Methodology, Supervision, Writing—review & editing.
Tengfei Yu: Investigation, Data interpretation.
Xin Yang: Conceptualization, Methodology, Data analysis.
Hongxia Zhang: Investigation, Data interpretation.
Jie Tian: Conceptualization, Project administration, Investigation, Supervision.
Wen He: Conceptualization, Project administration, Investigation, Supervision.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, G., Ning, B., Hui, H. et al. Image-to-Images Translation for Multiple Virtual Histological Staining of Unlabeled Human Carotid Atherosclerotic Tissue. Mol Imaging Biol 24, 31–41 (2022). https://doi.org/10.1007/s11307-021-01641-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-021-01641-w