Abstract
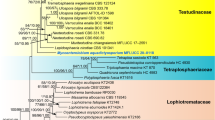
During surveys of ascomycetes from the Western Tien Shan Mountains in Uzbekistan, we collected a coelomycetous taxon on a dead trunk and branches of Lonicera altmannii. The taxon is morphologically distinct from other known melanommataceous taxa based on its unique set of characters, such as superficial to erumpent, uniloculate conidiomata, and globose ellipsoidal or ovoid, dark-brown, multiseptate conidia. It is introduced herein as Melanocamarosporioides ugamica gen. et sp. nov. within the family Melanommataceae. Multiloci phylogenies based on LSU, SSU, ITS, and TEF1-α sequence data generated from maximum likelihood, maximum parsimony and MrBayes analyses indicate that M. ugamica forms a distinct lineage within Melanommataceae and its establishment as a new genus is supported. Descriptions, illustrations, and appraisal on taxonomy for the taxa are provided.


Similar content being viewed by others
References
Abdel-Aziz FA (2016) Two new cheirosporous asexual taxa (Dictyosporiaceae, Pleosporales, Dothideomycetes) from freshwater habitats in Egypt. Mycosphere 7(4):448–457. https://doi.org/10.5943/mycosphere/7/4/5
Barr ME (1984) Herpotrichia and its segregates. Mycotaxon 20:1–38
Cai L, Jeewon R, Hyde KD (2006) Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology. Mycol Res 110:137–150
Castañeda RF, Heredia G, Reyes M, Arias RM, Decock C (2001) A revision of the genus Pseudospiropes and some new taxa. Cryptogam Mycol 22(1):3–18
Chesters CGC (1938) Studies on British pyrenomycetes II. A comparative study of Melanomma pulvis-pyrius (Pers.) Fuckel, Melanomma fuscidulum Sacc. and Thyridaria rubronotata (B.&Br.) Sacc. Trans Br Mycol Soc 22:116–150
Chomnunti P, Hongsanan S, Hudson BA, Tian Q, Peroh D, Dhami MK, Alias AS, Xu J, Liu X, Stadler M, Hyde KD (2014) The sooty moulds. Fungal Divers 66:1–36. https://doi.org/10.1007/s13225-014-0278-5
Crous PW, Summerell BA, Shivas RG, Romberg M, Mel’nik VA, Verkley GJM, Groenewald JZ (2011) Fungal planet description sheets: 92–106. Persoonia 27:130–162
Crous PW, Braun U, Hunter GC, Wingfield MJ, Verkley GJM, Shin HD, Nakashima C, Groenewald JZ (2013) Phylogenetic lineages in Pseudocercospora. Stud Mycol 75:37–114
Crous PW, Groenewald JZ (2017) The Genera of Fungi – G 4: Camarosporium and Dothiora. IMA Fungus 8(1):131–152
Gupta VK, Tuohy MG, Ayyachamy M, Turner KM, O’Donovan A (2013) Current methods in fungal biology. In: Laboratory protocols in fungal biology. Springer, London, pp 114–115. https://doi.org/10.1007/978-1-4614-2356-0
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser:95–98
Hashimoto A, Matsumura M, Hirayama K, Fujimoto R, Tanaka K (2017) Pseudodidymellaceae fam. nov.: phylogenetic affiliations of mycopappus-like genera in Dothideomycetes. Stud Mycol 87:187–206
Hernández-Restrepo M, Gené J, Castañeda-Ruiz RF, Mena-Portales J, Crous PW, Guarro J (2017) Phylogeny of saprobic microfungi from southern Europe. Stud Mycol 86:53–97. https://doi.org/10.1016/j.simyco.2017.05.002
Ho WH, Yanna, Hyde KD, Hodgkiss IJ (2002) Seasonality and sequential occurrence of fungi on wood submerged in Tai Po Kau Forest Stream, Hong Kong. Fungal Divers 10:21–43
Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310–2314
Hyde KD, Jones EBG, Liu JK et al (2013) Families of Dothideomycetes. Fungal Divers 63:1–313
Hyde KD, Hongsanan S, Jeewon R et al (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 78:1–237
Index Fungorum (2018) Index Fungorum. http://www.indexfungorum.org/names/Names.asp. Accessed Jan 2018
Jaklitsch WM, Voglmayr H (2017) Three former taxa of Cucurbitaria and considerations on Petrakia in the Melanommataceae. Sydowia 69:81–95
Jayasiri SC, Hyde KD, Ariyawansa HA et al (2015) The faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers 74(1):3–18
Jeewon R, Hyde KD (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7:1669–1677. https://doi.org/10.5943/mycosphere/7/11/4
Jeewon R, Liew ECY, Hyde KD (2002) Phylogenetic relationships of Pestalotiopsis and allied genera inferred from ribosomal DNA sequences and morphological characters. Mol Phylogenet Evol 25:378–392
Jeewon R, Liew ECY, Simpson JA, Hodgkiss IJ, Hyde KD (2003) Phylogenetic significance of morphological characters in the taxonomy of Pestalotiopsis species. Mol Phylogenet Evol 27:372–383
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Li WY, Zhuang WY (2008) Notes on the genus Byssosphaeria (Melanommataceae) from China. Mycosystema 27(1):48–53
Li J, Jeewon R, Luo Z, Phookamsak R, Bhat D, Mapook A, Phukhamsakda C, Camporesi E, Lumyong S, Hyde KD (2017) Morphological characterization and DNA based taxonomy of Fusiconidium gen. nov. with two novel taxa within Melanommataceae (Pleosporales). Phytotaxa 308(2):206–218 https://doi.org/10.11646/phytotaxa.308.2.2
Liew ECY, Aptroot A, Hyde KD (2003) Phylogenetic significance of the pseudoparaphyses in Loculoascomycete taxonomy. Mol Phylogenet Evol 16:392–402
Mugambi GK, Huhndorf SM (2009) Molecular phylogenetics of Pleosporales: Melanommataceae and Lophiostomataceae recircumscribed (Pleosporomycetidae, Dothideomycetes, Ascomycota). Stud Mycol 64:103–121
Nylander JAA (2004) MrModeltest 2.0. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala
Phookamsak R, Liu JK, McKenzie EHC, Manamgoda DS, Ariyawansa H, Thambugala KM, Dai DQ, Camporesi E, Chukeatirote E, Wijayawardene NN, Bahkali AH, Mortimer PE, Xu JC, Hyde KD (2014) Revision of Phaeosphaeriaceae. Fungal Divers 68:159–238
Rambaut A, Drummond A (2008) FigTree: tree figure drawing tool, version 1.2. 2. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh
Rannala B, Yang Z (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol 43:304–311
Rehner SA (2001) Primers for elongation factor 1-alpha (EF1-alpha). Available from: http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf. Accessed 22 Jan 2018
Saccardo PA (1880) Fungi Gallici ser. II. Michelia 2:39–135
Shearer CA, Raja HA, Miller AN, Nelson P, Tanaka K, Hirayama K, Marvanová L, Hyde KD, Zhang Y (2009) The molecular phylogeny of freshwater Dothideomycetes. Stud Mycol 64:145–153
Sivanesan A (1983) Studies on ascomycetes. Trans Br Mycol Soc 81:313–332
Sivanesan A (1984) The bitunicate ascomycetes and their anamorphs. J. Cramer. Vaduz, University of California. ISBN 3768213293
Su HY, Udayanga D, Luo ZL, Manamgoda DS, Zhao YC, Yang J, Liu XY, Mckenzie EHC, Zhou DQ, Hyde KD (2015) Hyphomycetes from aquatic habitats in Southern China: species of Curvularia (Pleosporaceae) and Phragmocephala (Melannomataceae). Phytotaxa 226(3):201–216
Sutton BC (1980) The coelomycetes. Fungi Imperfecti with Pycnidial acervuli and stromata. Commonwealth Mycological Institute, Kew, pp 1–696
Swofford DL (2002) PAUP: phylogenetic analysis using parsimony and other methods version 4.0 b10. Sinauer Associates, Sunderland
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tian Q, Liu JK, Hyde KD et al (2015) Phylogenetic relationships and morphological reappraisal of Melanommataceae (Pleosporales). Fungal Divers 74:267–324. https://doi.org/10.1007/s13225-015-0350-9
Tibpromma S, Hyde KD, Jeewon R, Maharachchikumbura SSN (2017) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 83(1):1–261
TreeBASE (2018) Retrieved from: www.treebase.org
Valenzuela-Lopez N, Cano-Lira JF, Guarro J, Sutton DA, Wiederhold N, Crous PW, Stchigel AM (2018) Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud Mycol 90:1–69. https://doi.org/10.1016/j.simyco.2017.11.003
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246
Wanasinghe DN, Hyde KD, Jeewon R, Crous PW, Wijayawardene NN, Jones EBG, Karunarathna SC (2017) Phylogenetic revision of Camarosporium (Pleosporineae, Dothideomycetes) and allied genera. Stud Mycol 87:207–256. https://doi.org/10.1016/j.simyco.2017.08.001
Wanasinghe DN, Phukhamsakda C, Hyde KD et al (2018) Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers 89:1–236
Wang HK, Aptroot A, Crous PW, Hyde KD, Jeewon R (2007) The polyphyletic nature of Pleosporales: an example from Massariosphaeria based on rDNA and RBP2 gene phylogenies. Mycol Res 111:1268–1276
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, San Diego, pp 315–322
Whitton SR, McKenzie EHC, Hyde KD (2012) Fungi associated with Pandanaceae. (Series editor: Hyde KD.), Fungal Divers Res Ser 21:1–457
Wijayawardene NN, Crous PW, Kirk PM et al (2014a) Naming and outline of Dothideomycetes—2014 including proposals for the protection or suppression of generic names. Fungal Divers 69:1–55
Wijayawardene NN, Hyde KD, Bhat DJ, Camporesi E, Schumacher RK, Chethana KWT, Wikee S, Bahkali AH, Wang Y (2014b) Camarosporium-like species are polyphyletic in Pleosporales; introducing Paracamarosporium and Pseudocamarosporium gen. nov. in Montagnulaceae. Cryptogam Mycol 35:177–198
Wijayawardene NN, Hyde KD, Wanasinghe DN et al (2016) Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers 77:1–316
Wijayawardene NN, Hyde KD, Rajeshkumar KC et al (2017) Notes for genera: Ascomycota. Fungal Divers 86(1):1–594
Wijayawardene NN, Hyde KD, Lumbsch TH, Liu JK, Maharachchikumbura SN, Ekanayaka AH, Tian Q, Phookamsak R (2018) Outline of Ascomycota: 2017. Fungal Divers 88(1):167–263
Winter G (1887) Ascomyceten. In: Rabenhorst’s DiePilzeDeutschlands, Oesterreichs und der Schweiz. Bd I, Abt II
Zhang Y, Wang HK, Fournier J, Crous PW, Jeewon R, Pointing SB, Hyde KD (2009) Towards a phylogenetic clarification of Lophiostoma/Massarina and morphologically similar genera in the Pleosporales. Fungal Divers 38:225–251
Zhang Y, Crous PW, Schoch CL, Hyde KD (2012) Pleosporales. Fungal Divers 53:1–221
Zhaxybayeva O, Gogarten JP (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3(1):1–15
Acknowledgements
We are grateful to Chiang Mai University and the Thailand Research Fund (TRF) grant no DBG6080013 entitled “The future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracaena species” and grant number RDG6130001 entitled “Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion.” The Mae Fah Luang University is thanked for supporting studies on Dothideomycetes fungi. Dhandevi Pem is also grateful to Qing Tian, Wei Dong, Deping Wei, Kasun Thambugala, Dhanushka Wanasinghe, Monika Dayarathne, Subashini Jayasiri, Rekhani Hansika Perera, and Milan C. Samarakoon for their precious help during this research. Yusufjon Gafforov acknowledges the Agency for Science and Technology of the Republic of Uzbekistan for financial support of research project (No. P3-2014-083017442, PЗ-2017-0921183) and the Chinese Academy of Sciences President’s International Fellowship Initiative for Visiting Scientist awarded (Grant No.: 2018VBB0021). We thank Nalin Wijayawardene for providing us with sequence data of Melanocamarosporium galiicola. The authors also thank Dr. Shaun Pennycook for nomenclatural review. Rajesh Jeewon thanks Mae Fah Luang University and University of Mauritius for research support.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Section Editor: Gerhard Rambold
Rights and permissions
About this article
Cite this article
Pem, D., Jeewon, R., Gafforov, Y. et al. Melanocamarosporioides ugamica gen. et sp. nov., a novel member of the family Melanommataceae from Uzbekistan. Mycol Progress 18, 471–481 (2019). https://doi.org/10.1007/s11557-018-1448-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-018-1448-8