Abstract
Purpose
Surra is a zoonotic disease caused by Trypanosoma evansi (Trypanozoon), a salivary trypanosome native to Africa which affects a wide range of mammals worldwide and causes mortality and significant economic loss. The present study was devoted to the molecular characterization of T. evansi derived from naturally infected dromedary camels in Algeria.
Methods
A total of 148 blood samples were collected from mixed age camels living in one of four geographic regions (Ouargla, El Oued, Biskra and Ghardaia) of Algeria. Samples underwent PCR amplification and sequencing of the internal transcribed spacer 1 (ITS1) complete sequence.
Results
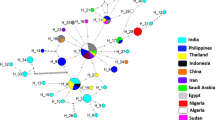
DNA of Trypanosoma spp. was found in 19 camels (12.84%). Trypanosoma spp. molecular positivity was not affected by sex (p = 0.50), age (p = 0.08), or geographic location (p = 0.12). Based on multiple sequence alignment of the obtained DNA sequences with representative T. evansi ITS1 sequences available globally, the Algerian sequences were grouped within four different haplotypes including two which were original.
Conclusion
Results of this study provide preliminary data on which future studies of genetic diversity and molecular epidemiology of T. evansi can be based.


Similar content being viewed by others
Availability of data and material
Data transparency.
References
Lai DH, Hashimi H, Lun ZR, Ayala FJ, Lukes J (2008) Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc Natl Acad Sci USA 105:1999–2004. https://doi.org/10.1073/pnas.0711799105
Desquesnes M, Dargantes A, Lai DH, Lun ZR, Holzmuller P, Jittapalapong S (2013) Trypanosoma evansi and surra: a review and perspectives on transmission, epidemiology and control, impact, and zoonotic aspects. Biomed Res Int 2013:321237. https://doi.org/10.1155/2013/321237
Nakayima J, Nakao R, Alhassan A, Mahama C, Afakye K, Sugimoto C (2012) Molecular epidemiological studies on animal trypanosomiases in Ghana. Parasit Vectors 5:217. https://doi.org/10.1186/1756-3305-5-217
Hoare CA (1965) Vampire bats as vectors and hosts of equine and bovine trypanosomes. Acta Trop 22:204–216. https://pubmed.ncbi.nlm.nih.gov/4379528
Amer S, Ryu O, Tada C, Fukuda Y, Inoue N, Nakai Y (2011) Molecular identification and phylogenetic analysis of Trypanosoma evansi from dromedary camels (Camelus dromedarius) in Egypt, a pilot study. Acta Trop 117:39–46. https://doi.org/10.1016/j.actatropica.2010.09.010
Salah AA, Robertson I, Mohamed A (2015) Estimating the economic impact of Trypanosoma evansi infection on production of camel herds in Somaliland. Trop Anim Health Prod 47:707–714. https://doi.org/10.1007/s11250-015-0780-0
Kamidi CM, Saarman NP, Dion K, Mireji PO, Ouma C, Murilla G, Aksoy S, Schnaufer A, Caccone A (2017) Multiple evolutionary origins of Trypanosoma evansi in Kenya. PLoS Negl Trop Dis 11:e0005895. https://doi.org/10.1371/journal.pntd.0005895
Aregawi WG, Agga GE, Abdi RD, Buscher P (2019) Systematic review and meta-analysis on the global distribution, host range, and prevalence of Trypanosoma evansi. Parasit Vectors 12:67. https://doi.org/10.1186/s13071-019-3311-4
Li Z, Pinto Torres JE, Goossens J, Stijlemans B, Sterckx YG, Magez S (2020) Development of a recombinase polymerase amplification lateral flow assay for the detection of active Trypanosoma evansi infections. PLoS Negl Trop Dis 14:e0008044. https://doi.org/10.1371/journal.pntd.0008044
Joshi PP, Shegokar VR, Powar RM, Herder S, Katti R, Salkar HR, Dani VS, Bhargava A, Jannin J, Truc P (2005) Human trypanosomiasis caused by Trypanosoma evansi in India: the first case report. Am J Trop Med Hyg 73:491–495. https://pubmed.ncbi.nlm.nih.gov/16172469.
Van Vinh CN, Buu CL, Desquesnes M, Herder S, Phu Huong LN, Campbell JI, Van CN, Yimming B, Chalermwong P, Jittapalapong S, Ramon FJ, Tri TN, Rabaa MA, Carrique-Mas J, Pham Thi TT, Tran Vu TN, Berto A, Thi HN, Van Minh HN, Canh TN, Khac CN, Wills B, Tinh HT, Thwaites GE, Yacoub S, Baker S (2016) A Clinical and Epidemiological Investigation of the First Reported Human Infection With the Zoonotic Parasite Trypanosoma evansi in Southeast Asia. Clin Infect Dis 62:1002–1008. https://doi.org/10.1093/cid/ciw052
Hassan-Kadle AA, Ibrahim AM, Nyingilili HS, Yusuf AA, Vieira TSWJ, Vieira RFC (2019) Parasitological, serological and molecular survey of camel trypanosomiasis in Somalia. Parasit Vectors 12:598. https://doi.org/10.1186/s13071-019-3853-5
Tehseen S, Jahan N, Qamar MF, Desquesnes M, Shahzad MI, Deborggraeve S, Buscher P (2015) Parasitological, serological and molecular survey of Trypanosoma evansi infection in dromedary camels from Cholistan Desert. Pakistan Parasit Vectors 8:415. https://doi.org/10.1186/s13071-015-1002-3
Chappuis F, Loutan L, Simarro P, Lejon V, Buscher P (2005) Options for field diagnosis of human African trypanosomiasis. Clin Microbiol Rev 18:133–146. https://doi.org/10.1128/CMR.18.1.133-146.2005
Masiga DK, Gibson WC (1990) Specific probes for Trypanosoma (Trypanozoon) evansi based on kinetoplast DNA minicircles. Mol Biochem Parasitol 40:279–283. https://doi.org/10.1016/0166-6851(90)90049-r
Birhanu H, Gebrehiwot T, Goddeeris BM, Buscher P, Van RN (2016) New Trypanosoma evansi Type B Isolates from Ethiopian Dromedary Camels. PLoS Negl Trop Dis 10:e0004556. https://doi.org/10.1371/journal.pntd.0004556
Ngaira JM, Njagi EN, Ngeranwa JJ, Olembo NK (2004) PCR amplification of RoTat 1.2 VSG gene in Trypanosoma evansi isolates in Kenya. Vet Parasitol 120:23–33. https://doi.org/10.1016/j.vetpar.2003.12.007
Songa BE, Hamers R (1988) A card agglutination test (CATT) for veterinary use based on an early VAT RoTat 1/2 of Trypanosoma evansi. Ann Soc Belg Med Trop 68: 233–240. https://pubmed.ncbi.nlm.nih.gov/3223785.
Geysen D, Delespaux V, Geerts S (2003) PCR-RFLP using Ssu-rDNA amplification as an easy method for species-specific diagnosis of Trypanosoma species in cattle. Vet Parasitol 110:171–180. https://doi.org/10.1016/s0304-4017(02)00313-8
da Silva FM, Noyes H, Campaner M, Junqueira AC, Coura JR, Anez N, Shaw JJ, Stevens JR, Teixeira MM (2004) Phylogeny, taxonomy and grouping of Trypanosoma rangeli isolates from man, triatomines and sylvatic mammals from widespread geographical origin based on SSU and ITS ribosomal sequences. Parasitology 129:549–561. https://doi.org/10.1017/s0031182004005931
Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC, Davila AM (2005) The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res 95:186–192. https://doi.org/10.1007/s00436-004-1267-5
Bellabidi M, Benaissa MH, Bissati-Bouafia S, Harrat Z, Brahmi K, Kernif T (2020) Coxiella burnetii in camels (Camelus dromedarius) from Algeria: Seroprevalence, molecular characterization, and ticks (Acari: Ixodidae) vectors. Acta Trop 206:105443. https://doi.org/10.1016/j.actatropica.2020.105443
Benfodil K, Ansel S, Mohamed-Cherif A, Ait-Oudhia K (2019) Prevalence of Trypanosoma evansi in horses (Equus caballus) and donkeys (Equus asinus) in El-Bayadh district, southwestern Algeria. J Hellenic Veterinary Med Soc 70:1631–1638. https://doi.org/10.12681/jhvms.21786
Benfodil K, Buscher P, Abdelli A, Van RN, Mohamed-Herif A, Ansel S, Fettata S, Dehou S, Bebronne N, Geerts M, Balharbi F, Ait-Oudhia K (2020) Comparison of serological and molecular tests for detection of Trypanosoma evansi in domestic animals from Ghardaia district. South Algeria Vet Parasitol 280:109089. https://doi.org/10.1016/j.vetpar.2020.109089
Bennoune O, Adili N, Amri K, Bennecib L, Ayachi A (2013) Trypanosomiasis of camels (Camelus dromedarius) in Algeria: first report. Veterinary Res Forum 4: 273–275. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4279620/pdf/vrf-4-273.pdf.
Boushaki D, Adel A, Dia ML, Buscher P, Madani H, Brihoum BA, Sadaoui H, Bouayed N, Kechemir IN (2019) Epidemiological investigations on Trypanosoma evansi infection in dromedary camels in the South of Algeria. Heliyon 5:e02086. https://doi.org/10.1016/j.heliyon.2019.e02086
Jaimes-DuenezJ T-C, Valencia-Hernandez A, Sanchez-Arevalo D, Poche-Ceballos A, Ortiz-Alvarez J, Mejia-Jaramillo AM (2017) Molecular diagnosis and phylogeographic analysis of Trypanosoma evansi in dogs (Canis lupus familiaris) suggest an epidemiological importance of this species in Colombia. Prev Veterinary Med 139:82–89. https://doi.org/10.1016/j.prevetmed.2017.02.007
Martinez-Flores WA, Palma-Garcia JM, Caballero-Ortega H, Del Viento-Camacho A, Lopez-Escamilla E, Martinez-Hernandez F, Vinuesa P, Correa D, Maravilla P (2017) Genotyping Toxoplasma gondii with the B1 gene in naturally infected sheep from an endemic region in the Pacific coast of Mexico. Vector Borne Zoonotic Dis 17:495–502. https://doi.org/10.1089/vbz.2016.2085
Drali R, Abi-Rached L, Boutellis A, Djossou F, Barker SC, Raoult D (2016) Host switching of human lice to new world monkeys in South America. Infect Genet Evol 39:225–231. https://doi.org/10.1016/j.meegid.2016.02.008
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Salim B, Bakheit MA, Kamau J, Nakamura I, Sugimoto C (2011) Molecular epidemiology of camel trypanosomiasis based on ITS1 rDNA and RoTat 1.2 VSG gene in the Sudan. Parasit Vectors 4:31. https://doi.org/10.1186/1756-3305-4-31
Nazem MR, Karimidehkordi M, Farhoodi Moghadam M (2020) Detection of Trypanosoma evansi in camel abortions (Camelus dromedarius) in Iran using polymerase chain reaction. Turkish J Veterinary Anim Sci 44: 1–6. http://journals.tubitak.gov.tr/veterinary/issues/vet-20-44-2/vet-44-2-25-1908-76.pdf.
Benaissa MH, Mimoune N, Bentria Y, Kernif T, Boukhelkhal A, Youngs CR, Kaidi R, Faye B, Halis Y (2020) Seroprevalence and risk factors for Trypanosoma evansi, the causative agent of surra in the dromedary camel (Camelus dromedarius) population in southeastern Algeria. Onderstepoort J Vet Res 87(1):a1891. https://doi.org/10.4102/ojvr.v87i1.1891
Khuchareontaworn S, Singhaphan P, Viseshakul N, Chansiri K (2007) Genetic diversity of Trypanosoma evansi in buffalo based on internal transcribed spacer (ITS) regions. J Vet Med Sci 69:487–493. https://doi.org/10.1292/jvms.69.487
Konnai S, Mekata H, Mingala CN, Abes NS, Gutierrez CA, Herrera JR, Dargantes AP, Witola WH, Cruz LC, Inoue N, Onuma M, Ohashi K (2009) Development and application of a quantitative real-time PCR for the diagnosis of Surra in water buffaloes. Infect Genet Evol 9:449–452. https://doi.org/10.1016/j.meegid.2009.01.006
Sudan V, Jaiswal AK, Shanker D, Verma AK (2017) First report of molecular characterization and phylogenetic analysis of RoTat 1.2 VSG of Trypanosoma evansi from equine isolate. Trop Anim Health Prod 49:1793–1796. https://doi.org/10.1007/s11250-017-1384-7
Claes F, Verloo D, De Waal DT, Majiwa PA, Baltz T, Goddeeris BM, Buscher P (2003) The expression of RoTat 1.2 variable surface glycoprotein (VSG) in Trypanosoma evansi and T. equiperdum. Vet Parasitol 116:209–216. https://doi.org/10.1016/s0304-4017(02)00359-x
Medkour H, Laidoudi Y, Lafri I, Davoust B, Mekroud A, Bitam I, Mediannikov O (2019) Canine vector-borne protozoa: molecular and serological investigation for Leishmania spp., Trypanosoma spp., Babesia spp., and Hepatozoon spp. in dogs from Northern Algeria. Vet Parasitol Reg Stud Rep 19:100353. https://doi.org/10.1016/j.vprsr.2019.100353
OIE (2019) Use of animals in research and education. In: OIE terrestrial animal health code. http://www.oie.int/index.php?id=169&L=0&htmfile=chapitre_aw_research_education
Acknowledgements
We thank Professor Curtis R. Youngs, Animal Science Department, Iowa State University, Ames, IA 50011, United States of America for improvement of our English. We are also grateful to Nour el houda Ammani, Amina Arib and Amira Azzouz for their participation in the study.
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
This study with camels was supervised by the division of biological resources of the Scientific and Technical Research Centre for Arid Areas (CRSTRA). It was conducted in accordance with the World Animal Health Organization (OIE) guiding principles on animal welfare included in the OIE Terrestrial Animal Health Code [40].Verbal consent of farm owners involved in this investigation was obtained prior to the collection of blood samples from their animals.
Rights and permissions
About this article
Cite this article
Boutellis, A., Bellabidi, M., Benaissa, M.H. et al. New Haplotypes of Trypanosoma evansi Identified in Dromedary Camels from Algeria. Acta Parasit. 66, 294–302 (2021). https://doi.org/10.1007/s11686-020-00316-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-020-00316-w