Abstract
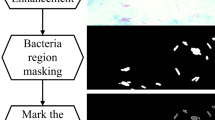
The World Health Organization suggests visual examination of stained sputum smear samples as a preliminary and basic diagnostic technique for diagnosing tuberculosis. The visual examination process requires much time of laboratorian, and also, it is prone to mistakes. For this purpose, this paper proposes a novel random forest (RF)-based segmentation and classification approaches for the automated classification of Mycobacterium tuberculosis in microscopic images of Ziehl–Neelsen-stained sputum smears obtained using a light-field microscope. The RF supervised learning method is improved to classify each pixel depending on local color distributions as a part of candidate bacilli regions. Therefore, each pixel is labeled as either a candidate tuberculosis (TB) bacilli pixel or not. The candidate pixels are grouped together using connected component analysis. Each pixel group is then rotated, resized and centrally positioned within a bounding box, respectively, in order to utilize appearance-based tuberculosis bacteria identification algorithms. Finally, each region is classified by using the proposed RF learning algorithm trained on manually marked TB bacteria regions in the training images. The algorithm produces results that agree well with manual segmentation and identification. Different two-class pixel and object classifiers are also compared to show the performance of the proposed RF-based pixel segmentation and bacilli objects identification algorithm. The sensitivity and specificity of the proposed classifier are above 75.77 and 96.97 % for the segmentation of the pixels, respectively. It is also revealed that the sensitivity increases over 93 % when the staining is performed in accordance with the procedure. Moreover, these measures are above 89.34 and 62.89 % for the identification of bacilli objects. The results show that the proposed novel method is quite successful when compared to the other applied methods.







Similar content being viewed by others
References
Palomino, J.C., Leao, S.C., Ritacco, V.: Tuberculosis 2007—From Basic Science to Patient Care. http://www.tuberculosistextbook.com/index.htm. Accessed June 2007
International Union Against Tuberculosis and Lung Disease: Sputum Examination for Tuberculosis by Direct Microscopy in Low Income Countries. France (2000)
Auramine-rhodamine Fluorescence-Acid Fast Bacteria. http://library.med.utah.edu/WebPath/webpath.html
Steingart, K., Hnery, M., Ng, V., Hopewell, P., Ramsay, A., Cunningham, J., Urbaczik, R., Perkins, M., Aziz, M., Pai, M.: Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect. Dis. 6(9), 570–581 (2006)
Revised National Tuberculosis Control Programme: Module for Laboratory Technicians. Central TB Division, New Delhi (2005)
Nguyen, T.N.L., Wells, C.D., Binkin, N.J., Pham, D.L., Nguyen, V.C.: The importance of quality control of sputum smear microscopy: the effect of reading errors on treatment decision and outcomes. Int. J. Tuberc. Lung Dis. 3(6), 483–487 (1999)
Forero, M.G., Sroubek, F., Cristobal, G.: Identification of tuberculosis bacteria based on shape and color. Real-Time Imaging 10, 251–262 (2004)
Forero, M.G., Cristobal, G.: Automatic identification techniques of tuberculosis bacteria. In: Proceedings of the SPIE, vol. 5203, pp. 71–81 (2003)
Forero, M.G., Cristobal, G., Desco, M.: Automatic identification of Mycobacterium tuberculosis by Gaussian mixture models. J. Microsc. 223, 120–132 (2006)
Veropoulos, K., Learmonth, G., Campbell, C., Knight, B., Simpson, J.: Automated identification of tubercle bacilli in sputum a preliminary investigation. Anal. Quant. Cytol. Histol. 21(4), 277–281 (1999)
Sadaphal, P., Rao, J., Comstock, G.W., Beg, M.F.: Image processing techniques for identifying Mycobacterium tuberculosis in Ziehl–Neelsen stains. Int. J. Tuberc. Lung Dis. 12(5), 579–582 (2008)
Siena, I., Adi, K., Gernowo, R., Mirnasari, N.: Development of algorithm tuberculosis bacteria identification using color segmentation and neural networks. Int. J. Video Image Process Netw. Secur. 12(4), 9–13 (2012)
Khutlang, R., Krishnan, S., Dendere, R., Whitelaw, A., Veropoulos, K., Learmonth, G., Douglas, T.S.: Classification of Mycobacterium tuberculosis in images of ZN-stained sputum smears. IEEE Trans. Inf. Technol. Biomed. 14(4), 949–957 (2010)
Alpaydn, E.: Introduction to Machine Learning. The MIT Press, London (2010)
Di Stefano, L., Bulgarelli, A.: A simple and efficient connected components labelling algorithm. In: International Conference on Image Analysis and Processing, pp. 322–327 (1999)
Hu, M.: Visual pattern recognition by moment invariants. IRE. Trans. Inf. Theor. 8, 179–187 (1962)
Do, C.B.: The multivariate Gaussian distribution. http://cs229.stanford.edu/section/gaussians (2008)
Cortes, C., Vapnik, V.: Support vector machines. Mach. Learn. 20, 273–297 (1995)
Breiman, L.: Bagging predictors. Mach. Learn. 24, 123–140 (1996)
Freund, Y., Schapire, R.E.: A short introduction to boosting. J. Jpn. Soc. Artif. Intell. 14(5), 771–780 (1999)
Ho, T.K.: The random subspace method for constructing decision forests. IEEE Trans. Pattern Anal. Mach. Intell. 20(8), 197–227 (1990)
Breiman, L.: Random forests. Mach. Learn. 45, 5–32 (2001)
Breiman, L., Friedman, J., Olshen, R., Stone, J.: Classification and Regression Trees. Chapman and Hall, Wadsworth, CA (1984)
Zurada, J.M.: Introduction to Artificial Neural Systems. West Publishing Co., St. Paul (1992)
KTU-CVPR Lab http://ceng2.ktu.edu.tr/cvpr/
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ayas, S., Ekinci, M. Random forest-based tuberculosis bacteria classification in images of ZN-stained sputum smear samples. SIViP 8 (Suppl 1), 49–61 (2014). https://doi.org/10.1007/s11760-014-0708-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-014-0708-6