Abstract
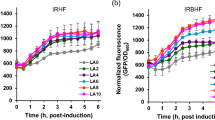
Glargine is a long-acting insulin analog with less hypoglycemia risk. Like human insulin, glargine is a globular protein composed of two polypeptide chains linked by two disulfide bonds. Pichia pastoris KM71 Muts strains were engineered to produce and secrete insulin glargine through the cleavage of two Kex2 sites. Nevertheless, the recombinant product was the single-chain insulin glargine (glargine precursor) instead of the expected double-chain glargine. Molecular model analysis of the dimeric and hexameric forms of the single-chain glargine showed buried Kex2 sites that prevent intracellular glargine precursor processing. The effect of the methanol-feeding strategy (methanol limited fed-batch vs. methanol non-limited fed-batch) and the induction temperature (28 °C vs. 24 °C) on the cell growth and production parameters in bioreactor cultures was also evaluated. Exponential growth at a constant specific growth rate was observed in all the cultures. The volumetric productivities and specific substrate consumption rates were directly proportional to the specific growth rate. The lower temperature led to increased metabolic activity of the yeast cells, which increased the specific growth rate. The methanol non-limited fed-batch culture at 24 °C showed the highest values for the process parameters. After 75 h of induction, 0.122 g/L of glargine precursor was obtained from the culture medium.





Similar content being viewed by others
Data Availability
Not applicable.
References
Saeedi, P., Petersohn, I., Salpea, P., Malanda, B., Karuranga, S., Unwin, N., & Shaw, J. E. (2019). Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas. Diabetes Research and Clinical Practice, 57, 107843. https://doi.org/10.1016/j.diabres.2019.107843.
Hilgenfeld, R., Seipke, G., Berchtold, H., & Owens, D. R. (2014). The evolution of insulin glargine and its continuing contribution to diabetes care. Drugs, 74, 911–927. https://doi.org/10.1007/s40265-014-0226-4.
Nagel, N., Graewert, M. A., Gao, M., Heyse, W., Jeffries, C. M., Svergun, D., & Berchtold, H. (2019). The quaternary structure of insulin glargine and glulisine under formulation conditions. Biophysical Chemistry, 253, 106226. https://doi.org/10.1016/j.bpc.2019.106226.
Potvin, G., Ahmad, A., & Zhang, Z. (2012). Bioprocess engineering aspects of heterologous protein production in Pichia pastoris: a review. Biochemical Engineering Journal, 64, 91–105. https://doi.org/10.1016/j.bej.2010.07.017.
Fuller, R. S., Sterne, R. E., & Thorner, J. (1988). Enzymes required for yeast prohormone processing. Annual Review of Physiology, 50, 345–362. https://doi.org/10.1146/annurev.ph.50.030188.002021.
Yang, S., Kuang, Y., Li, H., Liu, Y., Hui, X., Li, P., & Li, S. (2013). Enhanced production of recombinant secretory proteins in Pichia pastoris by optimizing Kex2 P1’ site. PLoS One, 8, e75347. https://doi.org/10.1371/journal.pone.0075347.
Rockwell, N. C., Krysan, D. J., Komiyama, T., & Fuller, R. S. (2002). Precursor processing by Kex2/furin proteases. Chemical Reviews, 102, 4525–4548. https://doi.org/10.1021/cr010168i.
Li, P., Anumanthan, A., Gao, X. G., Ilangovan, K., Suzara, V. V., Düzgüneş, N., & Renugopalakrishnan, V. (2007). Expression of recombinant proteins in Pichia pastoris. Applied Biochemistry and Biotechnology, 142, 105–124. https://doi.org/10.1007/s12010-007-0003-x.
País, J. M., Varas, L., Valdés, J., Cabello, C., Rodríguez, L., & Mansur, M. (2003). Modeling of mini-proinsulin production in Pichia pastoris using the AOX promoter. Biotechnology Letters, 25, 251–255. https://doi.org/10.1023/A:1022398917213.
País-Chanfrau, J. M., García, Y., Licor, L., Besada, V., Castellanos-Serra, L., Cabello, C. I., Hernández, L., Mansur, M., Plana, L., Hidalgo, A., Támbara, Y., Abrahantes-Pérez, M. C., Valdés, J., & Martínez, E. (2004). Improving the expression of mini-proinsulin in Pichia pastoris. Biotechnology Letters, 26, 1269–1272. https://doi.org/10.1023/B:BILE.0000044871.80632.f9.
Mansur, M., Cabello, C., Hernández, L., País, J., Varas, L., Valdés, J., Terrero, Y., Hidalgo, A., Plana, L., Besada, V., García, L., Lamazares, E., Castellanos, L., & Martínez, E. (2005). Multiple gene copy number enhances insulin precursor secretion in the yeast Pichia pastoris. Biotechnology Letters, 27, 339–345. https://doi.org/10.1007/s10529-005-1007-7.
Polez, S., Origi, D., Zahariev, S., Guarnaccia, C., Tisminetzky, S. G., Skoko, N., & Baralle, M. (2016). A simplified and efficient process for insulin production in Pichia pastoris. PLoS One, 11, e0167207. https://doi.org/10.1371/journal.pone.0167207.
Thongyoo, S., Phakham, T., Khongchareonporn, N., Reamtong, O., Karnchanatat, A., Phutong, S., Wichai, T., Noitang, S., & Sooksai, S. (2019). Expression, purification and biological activity of monomeric insulin precursors from methylotrophic yeasts. Protein Expression and Purification, 153, 35–43. https://doi.org/10.1016/j.pep.2018.08.002.
Cos, O., Ramón, R., Montesinos, J. L., & Valero, F. (2006). Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: a review. Microbial Cell Factories, 5, 17. https://doi.org/10.1186/1475-2859-5-17.
Looser, V., Bruhlmann, B., Bumbak, F., Stenger, C., Costa, M., Camattari, A., & Kovar, K. (2015). Cultivation strategies to enhance productivity of Pichia pastoris: a review. Biotechnology Advances, 33, 1177–1193. https://doi.org/10.1016/j.biotechadv.2015.05.008.
Barrigón, J. M., Montesinos, J. L., & Valero, F. (2013). Searching the best operational strategies for Rhizopus oryzae lipase production in Pichia pastoris Mut+ phenotype: methanol limited or methanol non-limited fed-batch cultures? Biochemical Engineering Journal, 75, 47–54. https://doi.org/10.1016/j.bej.2013.03.018.
Jahic, M., Gustavsson, M., Jansen, A. K., Martinelle, M., & Enfors, S. O. (2003). Analysis and control of proteolysis of a fusion protein in Pichia pastoris fed-batch processes. Journal of Biotechnology, 102, 45–53. https://doi.org/10.1016/S0168-1656(03)00003-8.
Dragosits, M., Stadlmann, J., Albiol, J., Baumann, K., Maurer, M., Gasser, B., Sauer, M., Altmann, F., Ferrer, P., & Mattanovich, D. (2009). The effect of temperature on the proteome of recombinant Pichia pastoris. Journal of Proteome Research, 8, 1380–1392. https://doi.org/10.1021/pr8007623.
Viader-Salvadó, J. M., Castillo-Galván, M., Fuentes-Garibay, J. A., Iracheta-Cárdenas, M. M., & Guerrero-Olazarán, M. (2013). Optimization of five environmental factors to increase beta-propeller phytase production in Pichia pastoris and impact on the physiological response of the host. Biotechnology Progress, 29, 1377–1385. https://doi.org/10.1002/btpr.1822.
Sreekrishna, K. (1993). Strategies for optimizing protein expression and secretion in the methylotrophic yeast Pichia pastoris. In R. H. Baltz, G. D. Hegeman, & P. L. Skatrud (Eds.), Industrial microorganisms: basic and applied molecular genetics (pp. 119–126). Washington, D.C: American Society for Microbiology.
Green, M. R., & Sambrook, J. (2012). Molecular cloning: a laboratory manual (fourth ed.). New York: Cold Spring Harbor Laboratory Press.
Viader-Salvadó, J. M., Fuentes-Garibay, J. A., Castillo-Galván, M., Iracheta-Cárdenas, M. M., Galán-Wong, L. J., & Guerrero-Olazarán, M. (2013). Shrimp (Litopenaeus vannamei) trypsinogen production in Pichia pastoris bioreactor cultures. Biotechnology Progress, 29, 11–16. https://doi.org/10.1002/btpr.1646.
Zhang, W., Inan, M., & Meagher, M. M. (2007). Rational design and optimization of fed-batch and continuous fermentations. In J. M. Cregg (Ed.), Methods in Molecular Biology: Pichia protocols (Second ed., pp. 43–64). New Jersey: Humana Press.
Katakura, Y., Zhang, W., Zhuang, G., Omasa, T., Kishimoto, M., Goto, Y., & Suga, K. I. (1998). Effect of methanol concentration on the production of human β2-glycoprotein I domain V by a recombinant Pichia pastoris: a simple system for the control of methanol concentration using a semiconductor gas sensor. Journal of Fermentation and Bioengineering, 86, 482–487. https://doi.org/10.1016/S0922-338X(98)80156-6.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research, 29, e45. https://doi.org/10.1093/nar/29.9.e45.
Waterhouse, A., Bertoni, M. B., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F. T., de Beer, T. A. P., Rempfer, C., Bordoli, L., Lepore, R., & Schwede, T. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research, 46, W296–W303. https://doi.org/10.1093/nar/gky427.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25, 1605–1612. https://doi.org/10.1002/jcc.20084.
Guex, N., & Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. https://doi.org/10.1002/elps.1150181505.
Laskowski, R. A., Jabłońska, J., Pravda, L., Vařeková, R. S., & Thornton, J. M. (2018). PDBsum: structural summaries of PDB entries. Protein Science, 27, 129–134. https://doi.org/10.1002/pro.3289.
Tress, M. (2013). Protein tertiary structures: prediction from amino acid sequences, in: eLS. Chichester: John Wiley & Sons, Ltd.
Xie, T., Liu, Q., Xie, F., Liu, H., & Zhang, Y. (2008). Secretory expression of insulin precursor in Pichia pastoris and simple procedure for producing recombinant human insulin. Preparative Biochemistry & Biotechnology, 38, 308–317. https://doi.org/10.1080/10826060802165147.
Gurramkonda, C., Polez, S., Skoko, N., Adnan, A., Gäbel, T., Chugh, D., Swaminathan, S., Khanna, N., Tisminetzky, S., & Rinas, U. (2010). Application of simple fed-batch technique to high-level secretory production of insulin precursor using Pichia pastoris with subsequent purification and conversion to human insulin. Microbial Cell Factories, 9, 31. https://doi.org/10.1186/1475-2859-9-31.
Sreenivas, S., Krishnaiah, S. M., Govindappa, N., Basavaraju, Y., Kanojia, K., Mallikarjun, N., & Sastry, K. N. (2015). Enhancement in production of recombinant two-chain insulin glargine by over-expression of Kex2 protease in Pichia pastoris. Applied Microbiology and Biotechnology, 99, 327–336. https://doi.org/10.1007/s00253-014-6052-5.
Wang, J., Wang, X., Shi, L., Zhang, Y., Zhou, X., & Cai, M. (2018). Reduced methanol input induces increased protein output by AOX1 promoter in a trans-acting elements engineered Pichia pastoris. Journal of Industrial Microbiology & Biotechnology, 45, 25–30. https://doi.org/10.1007/s10295-017-1988-y.
Viader-Salvadó, J. M., Gallegos-López, J. A., Carreón-Trevino, J. G., Castillo-Galván, M., Rojo-Domínguez, A., & Guerrero-Olazarán, M. (2010). Design of thermostable beta-propeller phytases with activity over a broad range of pHs and their overproduction by Pichia pastoris. Applied and Environmental Microbiology, 76, 6423–6430. https://doi.org/10.1128/AEM.00253-10.
Rockwell, N. C., & Thorner, J. W. (2004). The kindest cuts of all: crystal structures of Kex2 and furin reveal secrets of precursor processing. Trends in Biochemical Sciences, 29, 80–87. https://doi.org/10.1016/j.tibs.2003.12.006.
Fuentes-Garibay, J. A., Aguilar, C. N., Rodríguez-Herrera, R., Guerrero-Olazarán, M., & Viader-Salvadó, J. M. (2015). Tannase sequence from a xerophilic Aspergillus niger strain and production of the enzyme in Pichia pastoris. Molecular Biotechnology, 57, 439–447. https://doi.org/10.1007/s12033-014-9836-z.
Dodson, G., & Steiner, D. (1998). The role of assembly in insulin's biosynthesis. Current Opinion in Structural Biology, 8, 189–194. https://doi.org/10.1016/S0959-440X(98)80037-7.
Kjeldsen, T., & Pettersson, A. F. (2003). Relationship between self-association of insulin and its secretion efficiency in yeast. Protein Expression and Purification, 27, 331–337. https://doi.org/10.1016/S1046-5928(02)00640-X.
Müller II, F., Tieke, A., Waschk, D., Mühle, C., Müller, I. F., Seigelchifer, M., Pesce, A., Jenzelewskia, V., & Gellissen, G. (2002). Production of IFNα-2a in Hansenula polymorpha. Process Biochemistry, 38, 15–25. https://doi.org/10.1016/S0032-9592(02)00037-7.
Takahashi, S., Ueda, M., & Tanaka, A. (2000). Effect of the truncation of the C-terminal region of Kex2 endoprotease on processing of the recombinant Rhizopus oryzae lipase precursor in the co-expression system in yeast. Journal of Molecular Catalysis B: Enzymatic, 10, 233–240. https://doi.org/10.1016/S1381-1177(00)00130-2.
Brenner, C., & Fuller, R. S. (1992). Structural and enzymatic characterization of a purified prohormone-processing enzyme: secreted, soluble Kex2 protease. Proceedings of the National Academy of Sciences of the United States of America, 89, 922–926. https://doi.org/10.1073/pnas.89.3.922.
Aw, R., Barton, G. R., & Leak, D. J. (2017). Insights into the prevalence and underlying causes of clonal variation through transcriptomic analysis in Pichia pastoris. Applied Microbiology and Biotechnology, 101, 5045–5058. https://doi.org/10.1007/s00253-017-8317-2.
Viader-Salvadó, J. M., Cab-Barrera, E. L., Galán-Wong, L. J., & Guerrero-Olazarán, M. (2006). Genotyping of recombinant Pichia pastoris strains. Cellular & Molecular Biology Letters, 11, 348–359. https://doi.org/10.2478/s11658-006-0029-z.
Schwarzhans, J. P., Wibberg, D., Winkler, A., Luttermann, T., Kalinowski, J., & Friehs, K. (2016). Integration event induced changes in recombinant protein productivity in Pichia pastoris discovered by whole genome sequencing and derived vector optimization. Microbial Cell Factories, 15, 1–15. https://doi.org/10.1186/s12934-016-0486-7.
Vogl, T., Gebbie, L., Palfreyman, R. W., & Speight, R. (2018). Effect of plasmid design and type of integration event on recombinant protein expression in Pichia pastoris. Applied and Environmental Microbiology, 84, e02712–e02717. https://doi.org/10.1128/AEM.02712-17.
D'anjou, M. C., & Daugulis, A. J. (2001). A rational approach to improving productivity in recombinant Pichia pastoris fermentation. Biotechnology and Bioengineering, 72, 1–11. https://doi.org/10.1002/1097-0290(20010105)72:1<1::AID-BIT1>3.0.CO;2-T.
Charoenrat, T., Sangprapai, K., Promdonkoy, P., Kocharin, K., Tanapongpipat, S., & Roongsawang, N. (2015). Enhancement of thermostable β-glucosidase production in a slow methanol utilization strain of Pichia pastoris by optimization of the specific methanol supply rate. Biotechnology and Bioprocess Engineering, 20, 315–323. https://doi.org/10.1007/s12257-014-0686-0.
Potgieter, T. I., Kersey, S. D., Mallem, M. R., Nylen, A. C., & d'Anjou, M. (2010). Antibody expression kinetics in glycoengineered Pichia pastoris. Biotechnology and Bioengineering, 106, 918–927. https://doi.org/10.1002/bit.22756.
Hang, H. F., Chen, W., Guo, M. J., Chu, J., Zhuang, Y. P., & Zhang, S. (2008). A simple unstructured model-based control for efficient expression of recombinant porcine insulin precursor by Pichia pastoris. Korean Journal of Chemical Engineering, 25, 1065–1069. https://doi.org/10.1007/s11814-008-0174-3.
Brierley, R. A., Bussineau, C., Kosson, R., Melton, A., & Siegel, R. S. (1990). Fermentation development of recombinant Pichia pastoris expressing the heterologous gene: bovine lysozyme. Annals of the New York Academy of Sciences, 589, 350–362. https://doi.org/10.1111/j.1749-6632.1990.tb24257.x.
Vanz, A. L., Lünsdorf, H., Adnan, A., Nimtz, M., Gurramkonda, C., Khanna, N., & Rinas, U. (2012). Physiological response of Pichia pastoris GS115 to methanol-induced high level production of the Hepatitis B surface antigen: catabolic adaptation, stress responses, and autophagic processes. Microbial Cell Factories, 11, 103. https://doi.org/10.1186/1475-2859-11-103.
Zahrl, R., Mattanovich, D., & Gasser, B. (2018). The impact of ERAD on recombinant protein secretion in Pichia pastoris (syn Komagataella spp.). Microbiology (Reading, England), 164, 453–463. https://doi.org/10.1099/mic.0.000630.
Dietzsch, C., Spadiut, O., & Herwig, C. (2011). A dynamic method based on the specific substrate uptake rate to set up a feeding strategy for Pichia pastoris. Microbial Cell Factories, 10, 14. https://doi.org/10.1186/1475-2859-10-14.
Shay, L. K., Hunt, H. R., & Wegner, G. H. (1987). High-productivity fermentation process for cultivating industrial microorganisms. Journal of Industrial Microbiology, 2, 79–85. https://doi.org/10.1007/BF01569506.
Hwang, H. G., Kim, K. J., Lee, S. H., Kim, C. K., Min, C. K., Yun, J. M., & Son, Y. J. (2016). Recombinant glargine insulin production process using Escherichia coli. Journal of Microbiology and Biotechnology, 26, 1781–1789. https://doi.org/10.4014/jmb.1602.02053.
Acknowledgements
The authors thank Glen D. Wheeler for his stylistic suggestions in the preparation of the manuscript. A.C.-P. thanks Consejo Nacional de Ciencia y Tecnología (CONACYT) for his fellowship.
Funding
This work was supported by Universidad Autónoma de Nuevo León, Mexico (Grant number CN926-19, PAICYT).
Author information
Authors and Affiliations
Contributions
A.C.-P-: conceptualization, methodology, formal analysis, investigation, data curation, writing—original draft, writing—review and editing, visualization. J.M.V.-S.: conceptualization, methodology, formal analysis, data curation, writing—original draft, writing—review and editing, visualization, supervision, project administration. A.L.H.-E.: investigation, writing—original draft. J.A.F.-G.: investigation, data curation. M.G.-O.: conceptualization, methodology, resources, writing—review and editing, supervision, project administration, funding acquisition. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
This work does not involve experiments using humans or animals.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 14 kb)
Rights and permissions
About this article
Cite this article
Caballero-Pérez, A., Viader-Salvadó, J.M., Herrera-Estala, A.L. et al. Buried Kex2 Sites in Glargine Precursor Aggregates Prevent Its Intracellular Processing in Pichia pastoris Muts Strains and the Effect of Methanol-Feeding Strategy and Induction Temperature on Glargine Precursor Production Parameters. Appl Biochem Biotechnol 193, 2806–2829 (2021). https://doi.org/10.1007/s12010-021-03567-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03567-z