Abstract
We and others have previously demonstrated the potential for circulating exosome microRNAs to aid in disease diagnosis. In this study, we sought the possible utility of serum exosome microRNAs as biomarkers for disease activity in multiple sclerosis patients in response to fingolimod therapy. We studied patients with relapsing-remitting multiple sclerosis prior to and 6 months after treatment with fingolimod. Disease activity was determined using gadolinium-enhanced magnetic resonance imaging. Serum exosome microRNAs were profiled using next-generation sequencing. Data were analysed using univariate/multivariate modelling and machine learning to determine microRNA signatures with predictive utility. Accordingly, we identified 15 individual miRNAs that were differentially expressed in serum exosomes from post-treatment patients with active versus quiescent disease. The targets of these microRNAs clustered in ontologies related to the immune and nervous systems and signal transduction. While the power of individual microRNAs to predict disease status post-fingolimod was modest (average 77%, range 65 to 91%), several combinations of 2 or 3 miRNAs were able to distinguish active from quiescent disease with greater than 90% accuracy. Further stratification of patients identified additional microRNAs associated with stable remission, and a positive response to fingolimod in patients with active disease prior to treatment. Overall, these data underscore the value of serum exosome microRNA signatures as non-invasive biomarkers of disease in multiple sclerosis and suggest they may be used to predict response to fingolimod in future clinical practice. Additionally, these data suggest that fingolimod may have mechanisms of action beyond its known functions.





Similar content being viewed by others
Data Availability
The RNA-seq profile of exosomal miRNAs produced in this study is available at GEO repository with accession number GSE140106.
References
Milo R, Miller A (2014) Revised diagnostic criteria of multiple sclerosis. Autoimmun Rev 13(4–5):518–524
Thompson AJ et al (2018) Multiple sclerosis. Lancet 391(10130):14
Cree BA, Mares J, Hartung H-P (2019) Current therapeutic landscape in multiple sclerosis: an evolving treatment paradigm. 32(3):365–377
Ciccarelli OJTLN (2019) Multiple sclerosis in 2018: new therapies and biomarkers. 18(1):10–12
English C, Aloi JJ (2015) New FDA-approved disease-modifying therapies for multiple sclerosis. Clin Ther 37(4):691–715
Fonseca J (2015) Fingolimod real world experience: efficacy and safety in clinical practice
Chun J et al (2019) Fingolimod: lessons learned and new opportunities for treating multiple sclerosis and other disorders. 59:149–170
Ebrahimkhani S et al (2017) Exosomal microRNA signatures in multiple sclerosis reflect disease status. Sci Rep 7(1):14293
De Felice B et al (2014) Small non-coding RNA signature in multiple sclerosis patients after treatment with interferon-β. BMC Med Genet 7(1):26
Junker A, Hohlfeld R, Meinl E (2011) The emerging role of microRNAs in multiple sclerosis. Nat Rev Neurol 7(1):56–59
Fenoglio C et al (2016) Effect of fingolimod treatment on circulating miR-15b, miR23a and miR-223 levels in patients with multiple sclerosis. J Neuroimmunol 299:81–83
Waschbisch A et al (2011) Glatiramer acetate treatment normalizes deregulated microRNA expression in relapsing remitting multiple sclerosis. PLoS One 6(9):e24604
Gandhi R (2015) miRNA in multiple sclerosis: search for novel biomarkers. Mult Scler 21(9):1095–1103
Hecker M et al (2013) MicroRNA expression changes during interferon-beta treatment in the peripheral blood of multiple sclerosis patients. Int J Mol Sci 14(8):16087–16110
Ingwersen J et al (2015) Natalizumab restores aberrant miRNA expression profile in multiple sclerosis and reveals a critical role for miR-20b. Ann Clin Transl Neurol 2(1):43–55
Polman CH et al (2011) Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 69(2):292–302
Kurtzke JF (1983) Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology 33(11):1444–1444
Kaunzner UW, Gauthier SA (2017) MRI in the assessment and monitoring of multiple sclerosis: an update on best practice. Ther Adv Neurol Disord 10(6):247–261
Giorgio A, De Stefano N (2018) Effective utilization of MRI in the diagnosis and management of multiple sclerosis. Neurol Clin 36(1):27–34
Filippi M, Preziosa P, Rocca MA (2014) Magnetic resonance outcome measures in multiple sclerosis trials: time to rethink? Curr Opin Neurol 27(3):290–299
Filippi M et al (2019) Association between pathological and MRI findings in multiple sclerosis. 18(2):198–210
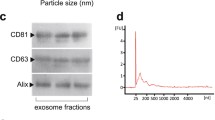
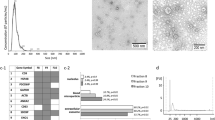
Lotvall J et al (2014) Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 3:26913
Charity W, Law YC, Shi W, Smyth GK (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29
Liu R et al (2015) Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res 43(15):e97
Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:3
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics:837–845
Tukey JW (1977) Exploratory data analysis. Vol. 2: Reading, Mass
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Liberzon A et al (2015) The molecular signatures database hallmark gene set collection. Cell Syst 1(6):417–425
Ru Y et al (2014) The multiMiR R package and database: integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res 42(17):e133–e133
Ludwig N et al (2016) Distribution of miRNA expression across human tissues. 44(8):3865–3877
Kappos L et al (2010) A placebo-controlled trial of oral fingolimod in relapsing multiple sclerosis. N Engl J Med 362(5):387–401
Kanehisa M et al (2016) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45(D1):D353–D361
Fabregat A et al (2017) The reactome pathway knowledgebase. Nucleic Acids Res 46(D1):D649–D655
Prager B, Spampinato SF, Ransohoff RM (2015) Sphingosine 1-phosphate signalling at the blood-brain barrier. Trends Mol Med 21(6):354–363
Pistono C et al (2017) What’s new about oral treatments in multiple sclerosis? Immunogenetics still under question. Pharmacol Res 120:279–293
Meira M et al (2014) Unravelling natalizumab effects on deregulated miR-17 expression in CD4+ T cells of patients with relapsing-remitting multiple sclerosis. J Immunol Res 2014:897249
Muñoz-Culla M, Irizar H, Castillo-Triviño T, Sáenz-Cuesta M, Sepúlveda L, Lopetegi I, de Munain AL, Olascoaga J et al (2014) Blood miRNA expression pattern is a possible risk marker for natalizumab-associated progressive multifocal leukoencephalopathy in multiple sclerosis patients. Mult Scler J 20(14):1851–1859
Guerau-de-Arellano M, Lovett-Racke AE, Racke MK (2010) miRNAs in multiple sclerosis: regulating the regulators. J Neuroimmunol 229(1–2):3–4
Yang Q, Pan W, Qian L (2017) Identification of the miRNA-mRNA regulatory network in multiple sclerosis. Neurol Res 39(2):142–151
Fenoglio C et al (2013) Decreased circulating miRNA levels in patients with primary progressive multiple sclerosis. Mult Scler J 19(14):1938–1942
De Santis G et al (2010) Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J Neuroimmunol 226(1–2):165–171
Zinger A et al (2016) Plasma levels of endothelial and B cell-derived microparticles are restored by fingolimod treatment in multiple sclerosis patients. Mult Scler J 22(14):1883–1887
Vistbakka J et al (2017) Circulating microRNAs as biomarkers in progressive multiple sclerosis. Mult Scler J 23(3):403–412
Husakova M (2016) MicroRNAs in the key events of systemic lupus erythematosus pathogenesis. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 160(3):327–342
Wu Y et al (2017) Lower serum levels of miR-29c-3p and miR-19b-3p as Biomarkers for Alzheimer’s disease. Tohoku J Exp Med 242(2):129–136
Rossi S et al (2014) Interleukin-1β causes excitotoxic neurodegeneration and multiple sclerosis disease progression by activating the apoptotic protein p53. Mol Neurodegener 9(1):56
Rawji KS, Yong VW (2013) The benefits and detriments of macrophages/microglia in models of multiple sclerosis. Clin Dev Immunol 2013:948976
Guarda G et al (2011) Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity 34(2):213–223
Bhinge A et al (2016) MiR-375 is essential for human spinal motor neuron development and may be involved in motor neuron degeneration. 34(1):124–134
Sievers C et al (2012) Altered microRNA expression in B lymphocytes in multiple sclerosis. Clin Immunol 144(1):70–79
Jernås M, Malmeström C, Axelsson M, Nookaew I, Wadenvik H, Lycke J, Olsson B (2013) MicroRNA regulate immune pathways in T-cells in multiple sclerosis (MS). BMC Immunol 14(32)
Martinelli-Boneschi F et al (2012) MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci Lett 508(1):4–8
Venken K et al (2008) Natural naive CD4+ CD25+ CD127low regulatory T cell (Treg) development and function are disturbed in multiple sclerosis patients: recovery of memory Treg homeostasis during disease progression. J Immunol 180(9):6411–6420
Severin ME et al (2016) MicroRNAs targeting TGFbeta signalling underlie the regulatory T cell defect in multiple sclerosis. Brain 139(Pt 6):1747–1761
Petrocca F, Vecchione A, Croce CM (2008) Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signalling. Cancer Res 68(20):8191–8194
Jana M et al (2014) Interleukin-12 (IL-12), but not IL-23, induces the expression of IL-7 in microglia and macrophages: implications for multiple sclerosis. Immunology 141(4):549–563
Libro R, Bramanti P, Mazzon E (2016) The role of the Wnt canonical signalling in neurodegenerative diseases. Life Sci 158:78–88
Acknowledgements
We would like to acknowledge and thank the patients who were involved in the study. We would like to thank the University of Sydney MS Clinical Trials staff for their assistance with venepuncture, serum preparation and storage and general study facilitation.
Funding
This project was partly funded by Novartis Australia.
Author information
Authors and Affiliations
Contributions
MHB, HNB, and MEB conceptualized and designed the study and provided critical review of the manuscript. HNB collected patients’ specimens, clinical and MRI information. SE and CW were involved in technical work for data acquisition; SE performed exosome purification and RNA extraction. FV developed the machine learning and bioinformatics pipelines. FV and SE analysed the data and generated the results. All the authors were involved in data interpretation. FV, SE and CMS drafted the manuscript and prepared the figures. All the authors revised and approved the final version.
Corresponding author
Ethics declarations
Ethical approval for the study was obtained from the University of Sydney Human Research Ethics Committee (2014/054).
Conflict of Interest
Heidi N Beadnall has received honoraria for presentations and advisory boards and has received educational travel support from Novartis, whom markets Gilenya®. Michael Barnett has received institutional support for research, speaking and/or participation in advisory boards for Biogen, Merck, Novartis, Roche and Sanofi Genzyme. He is a research consultant at Medical Safety Systems and research director for the Sydney Neuroimaging Analysis Centre. Michael E Buckland has received honoraria for presentations from Novartis and Biogen.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Saeideh Ebrahimkhani and Heidi N. Beadnall are equal authorship contribution.
Michael E. Buckland and Fatemeh Vafaee are equal authorship contribution.
Rights and permissions
About this article
Cite this article
Ebrahimkhani, S., Beadnall, H.N., Wang, C. et al. Serum Exosome MicroRNAs Predict Multiple Sclerosis Disease Activity after Fingolimod Treatment. Mol Neurobiol 57, 1245–1258 (2020). https://doi.org/10.1007/s12035-019-01792-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-01792-6