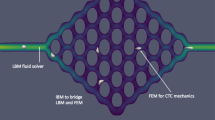
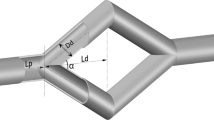
Abstract
Introduction
The biological and mechanical properties of circulating tumor cells (CTCs) in combination with the hemodynamics affect the preference of metastatic sites in the vasculature. Despite the extensive literature on the effects of biological properties on cell adhesion, the effects of hydrodynamic forces on primary attachment remains an active area of research. Using simulations in conjunction with experimentation, we provide new insight into the interplay of CTCs dynamics and local hydrodynamics.
Methods
A flow experiment of CTC attachment was performed within a bioprinted, double branching endothelialized vessel. Simulations of fluid flow and CTC transport in the reconstructed and idealized bifurcated vessel were respectively performed by HARVEY, our in-house massively parallel computational fluid dynamics solver. HARVEY is based on the lattice Boltzmann and finite element methods to model the fluid and cells dynamics. The immersed boundary method is employed for resolving the fluid–structure interaction.
Results
CTC attachment was quantified experimentally at all regions of the complex vessel. The results demonstrate a clear preference for CTCs to attach at the branch points. To elucidate the effect of the vessel topology on the location of attachment, a fluid-only simulation was performed assessing the differences in the hydrodynamics along the vessel. CTC transport in idealized bifurcated vessels was subsequently studied to examine the effects of cell deformability on the local hydrodynamics patterns and, thus, the preference of attachment sites.
Conclusions
The current work provides evidence on the correlation of the hydrodynamics forces arising from the vessel topology and CTC properties on the attachment regions.









Similar content being viewed by others
References
Anderson, K. J., A. de Guillebon, A. D. Hughes, W. Wang, and M. R. King. Effect of circulating tumor cell aggregate configuration on hemodynamic transport and wall contact. Math. Biosci. 294, 181–194, 2017.
Balogh, P., and P. Bagchi. Analysis of red blood cell partitioning at bifurcations in simulated microvascular networks. Phys. Fluids 30:051902, 2018
Barber, J. O., J. P. Alberding, J. M. Restrepo, and T. W. Secomb. Simulated two-dimensional red blood cell motion, deformation and partitioning in microvessel bifurcations. Ann. Biomed. Eng. 36:1690–1698, 2008.
Bhatnagar, P. L., E. P. Gross, and M. Krook. A model for collision processes in gases. I. Small amplitude processes in charged and neutral one-component systems. Phys. Rev. 94:511–525, 1954.
Cirak, F., M. Ortiz, and P. Schröder. Subdivision surfaces: a new paradigm for thin-shell finite-element analysis. Int. J. Numer. Methods Eng. 47:2039–2072, 2000.
Correia Faria, E., N. Ma, E. Gazi, P. Gardner, M. Brown, N. W. Clarke, and R. D. Snook. Measurement of elastic properties of prostate cancer cells using AFM. Analyst 133:1498–1500, 2008.
Dabagh, M., J. Gounley, and A. Randles. Localization of rolling and firm–adhesive interactions between circulating tumor cells and the microvasculature wall. Cell. Mol. Bioeng. 13:141–154, 2020.
Dabagh, M., and A. Randles. Role of deformable cancer cells on wall shear stress-associated-VEGF secretion by endothelium in microvasculature. PLoS ONE 14:e0211418, 2019.
dela Paz, N. G., T. E. Walshe, L. L. Leach, M. Saint-Geniez, and P. A. D’Amore. Role of shear-stress-induced VEGF expression in endothelial cell survival. J. Cell Sci. 125:831–843, 2012.
Dong, C., J. Cao, E. J. Struble, and H. H. Lipowsky. Mechanics of leukocyte deformation and adhesion to endothelium in shear flow. Ann. Biomed. Eng. 27:298–312, 1999.
Dong, C., and X. X. Lei. Biomechanics of cell rolling: shear flow, cell–surface adhesion, and cell deformability. J. Biomech. 33:35–43, 2000.
Doyeux, V., T. Podgorski, S. Peponas, M. Ismail, and G. Coupier. Spheres in the vicinity of a bifurcation: elucidating the Zweifach–Fung effect. J. Fluid Mech. 674:359–388, 2011.
Feiger, B., M. Vardhan, J. Gounley, M. Mortensen, P. Nair, R. Chaudhury, D. Frakes, and A. Randles. Suitability of lattice Boltzmann inlet and outlet boundary conditions for simulating flow in image-derived vasculature. Int. J. Numer. Methods Biomed. Eng. 35:e3198, 2019.
Follain, G., N. Osmani, A. S. Azevedo, G. Allio, L. Mercier, M. A. Karreman, G. Solecki, M. J. Garcia Lèon, O. Lefebvre, N. Fekonja, C. Hille, V. Chabannes, G. Dollé, T. Metivet, F. Der Hovsepian, C. Prudhomme, A. Pichot, N. Paul, R. Carapito, S. Bahram, B. Ruthensteiner, A. Kemmling, S. Siemonsen, T. Schneider, J. Fiehler, M. Glatzel, F. Winkler, Y. Schwab, K. Pantel, S. Harlepp, and J. G. Goetz. Hemodynamic forces tune the arrest, adhesion and extravasation of circulating tumor cells. Dev. Cell 45:33–52, 2018.
Gounley, J., E. W. Draeger, and A. Randles. Numerical simulation of a compound capsule in a constricted microchannel. Procedia Comput. Sci. 108, 175–184 (2017). In: International Conference on Computational Science, ICCS 2017, 12–14 June 2017, Zurich, Switzerland.
Green, A. E., and J. E. Adkins. Large Elastic Deformations. Oxford: Oxford University Press, 1960.
Guo, P., B. Cai, M. Lei, Y. Liu, and B. M. Fu. Differential arrest and adhesion of tumor cells and microbeads in the microvasculature. Biomech. Model. Mechanobiol. 13:537–550, 2014.
Guo, Z., C. Zheng, and B. Shi. Discrete lattice effects on the forcing term in the lattice Boltzmann method. Phys. Rev. E 65:046308, 2002.
Häner, E., M. Heil, and A. Juel. Deformation and sorting of capsules in a T-junction. J. Fluid Mech. 885:A4, 2020.
He, X., Q. Zou, L. S. Luo, and M. Dembo. Analytic solutions of simple flows and analysis of nonslip boundary conditions for the lattice Boltzmann BGK model. J. Stat. Phys. 87:115–136, 1997.
Hecht, M., and J. Harting. Implementation of on-site velocity boundary conditions for D3Q19 lattice Boltzmann simulations. J. Stat. Mech. 2010:P01018, 2010.
Huang, Q., X. Hu, W. He, Y. Zhao, S. Hao, Q. Wu, S. Li, S. Zhang, and M. Shi. Fluid shear stress and tumor metastasis. Am. J. Cancer Res. 8:763–777, 2018.
Hyakutake, T., and S. Nagai. Numerical simulation of red blood cell distributions in three-dimensional microvascular bifurcations. Microvasc. Res. 97:115–123, 2015.
Jadhav, S., C. D. Eggleton, and K. Konstantopoulos. A 3-D computational model predicts that cell deformation affects selectin-mediated leukocyte rolling. Biophys. J. 88:96–104, 2005.
Kaliviotis, E., J. M. Sherwood, and S. Balabani. Partitioning of red blood cell aggregates in bifurcating microscale flows. Sci. Rep. 7:44563, 2017.
Khismatullin, D. B., and G. A. Truskey. A 3D numerical study of the effect of channel height on leukocyte deformation and adhesion in parallel-plate flow chambers. Microvasc. Res. 68:188–202, 2004.
King, M. R., K. G. Phillips, A. Mitrugno, T. R. Lee, A. M. E. de Guillebon, S. Chandrasekaran, M. J. McGuire, R. T. Carr, S. M. Baker-Groberg, R. A. Rigg, A. Kolatkar, M. Luttgen, K. Bethel, P. Kuhn, P. Decuzzi, and O. J. T. McCarty. A physical sciences network characterization of circulating tumor cell aggregate transport. Am. J. Physiol. Cell Physiol. 308:C792–C802, 2015.
Kolesky, D. B., K. A. Homan, M. A. Skylar-Scott, and J. A. Lewis. Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl Acad. Sci. USA 113:3179–3184, 2016.
Krüger, T., F. Varnik, and D. Raabe. Efficient and accurate simulations of deformable particles immersed in a fluid using a combined immersed boundary lattice Boltzmann finite element method. Comput. Math. Appl. 61:3485–3505, 2011.
Lawrence, M. B., L. V. McIntire, and S. G. Eskin. Effect of flow on polymorphonuclear leukocyte/endothelial cell adhesion. Blood 70:1284–1290, 1987.
Lawrence, M. B., C. W. Smith, S. G. Eskin, and L. V. McIntire. Effect of venous shear stress on CD18-mediated neutrophil adhesion to cultured endothelium. Blood 75:227–237, 1990.
Leble, V., R. Lima, R. Dias, C. Fernandes, T. Ishikawa, Y. Imai, and T. Yamaguchi. Asymmetry of red blood cell motions in a microchannel with a diverging and converging bifurcation. Biomicrofluidics 5:044120, 2011.
Lekka, M., K. Pogoda, J. Gostek, O. Klymenko, S. Prauzner-Bechcicki, J. Wiltowska-Zuber, J. Jaczewska, J. Lekki, and Z. Stachura. Cancer cell recognition—mechanical phenotype. Micron 43:1259–1266, 2012.
Li, Q. S., G. Y. H. Lee, C. N. Ong, and C. T. Lim. AFM indentation study of breast cancer cells. Biochem. Biophys. Res. Commun. 374:609–613, 2008.
Loop, C. Smooth Subdivision Surfaces Based on Triangles. Master’s Thesis, The University of Utah, 1987.
Melder, R. J., L. L. Munn, S. Yamada, C. Ohkubo, and R. K. Jain. Selectin- and integrin-mediated T-lymphocyte rolling and arrest on TNF-\(\alpha \)-activated endothelium: augmentation by erythrocytes. Biophys. J. 69:2131–2138, 1995.
Munn, L. L., R. J. Melder, and R. K. Jain. Role of erythrocytes in leukocyte–endothelial interactions: mathematical model and experimental validation. Biophys. J. 71:466–478, 1996.
Peskin, C. S. The immersed boundary method. Acta Numer. 11:479–517, 2002.
Phillips, K. G., A. M. Lee, G. W. Tormoen, R. A. Rigg, A. Kolatkar, M. Luttgen, K. Bethel, L. Bazhenova, P. Kuhn, P. Newton, and O. J. T. McCarty. The thrombotic potential of circulating tumor microemboli: computational modelling of circulating tumor cell-induced coagulation. Am. J. Physiol. Cell Physiol. 308:C229–C236, 2015.
Pulaski, B. A., and S. Ostrand-Rosenberg. Mouse 4T1 breast tumor model. Curr. Protoc. Immunol. 39:20.2.1–20.2.16, 2000.
Qian, Y. H., D. D’Humières, and P. Lallemand. Lattice BGK models for Navier–Stokes equation. EPL 17:479–484, 1992.
Randles, A. P., V. Kale, J. Hammond, W. Gropp, and E. Kaxiras. Performance analysis of the lattice Boltzmann model beyond Navier–Stokes. In: 2013 IEEE 27th International Symposium on Parallel and Distributed Processing (IPDPS). IEEE, 2013, pp. 1063–1074.
Regmi, S., A. Fu, and K. Q. Luo. High shear stresses under exercise condition destroy circulating tumor cells in a microfluidic system. Sci. Rep. 7:39975, 2017.
Roychowdhury, S., J. Gounley, and A. Randles. Evaluating the influence of hemorheological parameters on circulating tumor cell trajectory and simulation time. In: The Platform for Advanced Scientific Computing (PASC) Conference. ACM, 2020.
Secomb, T. W., B. Styp-Rekowska, and A. R. Pries. Two-dimensional simulation of red blood cell deformation and lateral migration in microvessels. Ann. Biomed. Eng. 35:755–765, 2007.
Skalak, R., A. Tozeren, R. P. Zarda, and S. Chien. Strain energy function of red blood cell membranes. Biophys. J. 13:245–264, 1973.
Takeishi, N., Y. Imai, T. Yamaguchi, and T. Ishikawa. Flow of a circulating tumor cell and red blood cells in microvessels. Phys. Rev. E 92:063011, 2015.
Towns, J., T. Cockerill, M. Dahan, I. Foster, K. Gaither, A. Grimshaw, V. Hazlewood, S. Lathrop, D. Lifka, G. D. Peterson, et al. Xsede: accelerating scientific discovery. Comput. Sci. Eng. 16(5):62–74, 2014.
Urbich, C., M. Stein, K. Reisinger, R. Kaufmann, S. Dimmeler, and J. Gille. Fluid shear stress-induced transcriptional activation of the vascular endothelial growth factor receptor-2 gene requires Sp1-dependent DNA binding. FEBS Lett. 535:87–93, 2003.
Wang, Z., Y. Sui, A. V. Salsac, D. Barthès-Biesel, and W. Wang. Motion of a spherical capsule in branched tube flow with finite inertia. J. Fluid Mech. 806:603–626, 2016.
Wang, Z., Y. Sui, A. V. Salsac, D. Barthès-Biesel, and W. Wang. Path selection of a spherical capsule in a microfluidic branched channel: towards the design of an enrichment device. J. Fluid Mech. 849:136–162, 2018.
Wirtz, D., K. Konstantopoulos, and P. C. Searson. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer 11:512–522, 2011.
Woolfenden, H. C., and M. G. Blyth. Motion of a two-dimensional elastic capsule in a branching channel flow. J. Fluid Mech. 669:3–31, 2011.
Wu, P. H., D. Raz-Ben Aroush, A. Asnacios, W. C. Chen, M. E. Dokukin, B. L. Doss, P. Durand-Smet, A. Ekpenyong, J. Guck, N. V. Guz, P. A. Janmey, J. S. H. Lee, N. M. Moore, A. Ott, Y. C. Poh, R. Ros, M. Sander, I. Sokolov, J. R. Staunton, N. Wang, G. Whyte, and D. Wirtz. A comparison of methods to assess cell mechanical properties. Nat. Methods 15:491–498, 2018.
Xiao, L. L., Y. Liu, S. Chen, and B. M. Fu. Effects of flowing RBCs on adhesion of a circulating tumor cell in microvessels. Biomech. Model. Mechanobiol. 16:597–610, 2017.
Xiong, W., and J. Zhang. Two-dimensional lattice Boltzmann study of red blood cell motion through microvascular bifurcation: cell deformability and suspending viscosity effects. Biomech. Model. Mechanobiol. 11:575–583, 2012.
Xu, Y., F. Tian, H. Li, and Y. Deng. Red blood cell partitioning and blood flux redistribution in microvascular bifurcation. Theor. Appl. Mech. Lett. 2:024001, 2012.
Yan, W. W., B. Cai, Y. Liu, and B. M. Fu. Effects of wall shear stress and its gradient on tumor cell adhesion in curved microvessels. Biomech. Model. Mechanobiol. 11:641–653, 2012.
Yan, W. W., Y. Liu, and B. M. Fu. Effects of curvature and cell–cell interaction on cell adhesion in microvessels. Biomech. Model. Mechanobiol. 9:629–640, 2010.
Ye, H., H. Huang, and X. Lu. Numerical study on dynamic sorting of a compliant capsule with a thin shell. Comput. Fluids 114:110–120, 2015.
Yin, X., T. Thomas, and J. Zhang. Multiple red blood cell flows through microvascular bifurcations: cell free layer, cell trajectory, and hematocrit separation. Microvasc. Res. 89:47–56, 2013.
Acknowledgments
Research reported in this publication was supported by the Office of the Director of the National Institutes of Health under Award Number DP5OD019876. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was funded by LDRD 17ERD054 and LDRD 18ERD062 and performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344 (LLNL-JRNL-805606). Computing support for this work came from the LLNL Institutional Computing Grand Challenge Program. This work also used the Extreme Science and Engineering Discovery Environment (XSEDE) resource, Stampede2, at the Texas Advanced Computing Center through Allocation TG-IBN190011.48 The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that have contributed to the research results reported within this paper.
Conflict of interest
Marianna Pepona, Peter Balogh, Daniel F. Puleri, William F. Hynes, Claire Robertson, Karen Dubbin, Javier Alvarado, Monica L. Moya, and Amanda Randles declare that they have no conflict of interest.
Ethical Approval
No human studies were carried out by the authors for this article. No animal studies were carried out by the authors for this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Scott Simon oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pepona, M., Balogh, P., Puleri, D.F. et al. Investigating the Interaction Between Circulating Tumor Cells and Local Hydrodynamics via Experiment and Simulations. Cel. Mol. Bioeng. 13, 527–540 (2020). https://doi.org/10.1007/s12195-020-00656-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-020-00656-7