Abstract
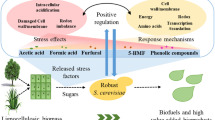
Efficient use of lignocellulosic biomass is a prerequisite for sustainable biofuel production while simultaneously contributing to environmental protection. However, phenolic compounds produced during the chemical treatment of lignocellulose inhibit the growth and metabolism of microorganisms, such as Escherichia coli, which is one of the ideal strains for producing target products by microbial fermentation. To provide new ideas for studying microbial tolerance to environmental stress and providing technical support for constructing the engineering strains with high yields of phenolic compounds, this review elucidates the inhibition mechanism of phenols to E. coli. Secondly, a comprehensive and systematic review of current approaches for improving the phenolic-tolerance of E. coli is provided, including strain domestication, random mutagenesis, regulating the expression of outer membrane proteins, changing the composition of membrane fatty acids, accelerating the efflux of phenolic compounds, engineered bacterial biofloc formation, and transcriptome analysis. Finally, this review ends with some questions that still exist today, and prospective strategies are outlined for further improving the phenols-tolerance of E. coli. This review provided a theoretical basis for research into microbial tolerance to environmental stress and the development of engineered strains with high yield of phenolic compounds.
Similar content being viewed by others
References
Bartolomé, B., M. L. Bengoechea, M. C. Gálvez, F. J. Pérez-Ilzarbe, T. Hernández, I. Estrella, and C. Gómez-Cordovés (1993) Photodiode array detection for elucidation of the structure of phenolic compounds. J. Chromatogr. A. 655: 119–125.
Cheniany, M., H. Ebrahimzadeh, A. Masoudi-nejad, K. Vahdati, and C. Leslie (2010) Effect of endogenous phenols and some antioxidant enzyme activities on rooting of Persian walnut (Juglans regia L.). Afr. J. Plant Sci. 4: 479–487.
Wang, Q., K. Zhou, Y. Ning, and G. Zhao (2016) Effect of the structure of gallic acid and its derivatives on their interaction with plant ferritin. Food Chem. 213: 260–267.
Wang, J., X. Shen, J. Rey, Q. Yuan, and Y. Yan (2018) Recent advances in microbial production of aromatic natural products and their derivatives. Appl. Microbiol. Biotechnol. 102: 47–61.
Limpisophon, K. and G. Schleining (2017) Use of gallic acid to enhance the antioxidant and mechanical properties of active fish gelatin film. J. Food Sci. 82: 80–89.
Chasov, A. V. and F. V. Minibayeva (2009) Effect of exogenous phenols on superoxide production by extracellular peroxidase from wheat seedling roots. Biochemistry (Mosc.) 74: 766–774.
Cao, B., K. Nagarajan, and K. C. Loh (2009) Biodegradation of aromatic compounds: current status and opportunities for biomolecular approaches. Appl. Microbiol. Biotechnol. 85: 207–228.
Liao, J. C., L. Mi, S. Pontrelli, and S. Luo (2016) Fuelling the future: microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 14: 288–304.
Peralta-Yahya, P. P., F. Zhang, S. B. del Cardayre, and J. D. Keasling (2012) Microbial engineering for the production of advanced biofuels. Nature. 488: 320–328.
Kakar, R., T. S. M. Amelia, C. C. Teng, K. Bhubalan, and A.-A. A. Amirul (2022) Biotransformation of oleochemical industry by-products to polyhydroxyalkanoate bioplastic using microbial fermentation: a review. Environ. Qual. Manag. 31: 31–46.
Liu, L., X. Duan, and J. Wu (2016) L-tryptophan production in Escherichia coli improved by weakening the pta-AckA pathway. PLoS One. 11: e0158200.
Liu, L., M. Bilal, X. Duan, and H. M. N. Iqbal (2019) Mitigation of environmental pollution by genetically engineered bacteria — current challenges and future perspectives. Sci. Total Environ. 667: 444–454.
Gu, H., J. Zhang, and J. Bao (2015) High tolerance and physiological mechanism of Zymomonas mobilis to phenolic inhibitors in ethanol fermentation of corncob residue. Biotechnol. Bioeng. 112: 1770–1782.
Liu, J., Q. Lin, X. Chai, Y. Luo, and T. Guo (2018) Enhanced phenolic compounds tolerance response of Clostridium beijerinckii NCIMB 8052 by inactivation of Cbei_3304. Microb. Cell Fact. 17: 35.
Luo, H., Z. Liu, F. Xie, M. Bilal, and F. Peng (2021) Lignocellulosic biomass to biobutanol: toxic effects and response mechanism of the combined stress of lignin-derived phenolic acids and phenolic aldehydes to Clostridium acetobutylicum. Ind. Crops Prod. 170: 113722.
Pontrelli, S., T. Y. Chiu, E. I. Lan, F. Y. Chen, P. Chang, and J. C. Liao (2018) Escherichia coli as a host for metabolic engineering. Metab. Eng. 50: 16–46.
Shin, J., Y. S. Jin, Y. C. Park, J. B. Park, Y. O. Lee, S. K. Kim, and D. H. Kweon (2021) Enhancing acid tolerance of Escherichia coli via viroporin-mediated export of protons and its application for efficient whole-cell biotransformation. Metab. Eng. 67: 277–284.
Zhu, T., D. Yao, D. Li, H. Xu, S. Jia, C. Bi, J. Cai, X. Zhu, and X. Zhang (2021) Multiple strategies for metabolic engineering of Escherichia coli for efficient production of glycolate. Biotechnol. Bioeng. 118: 4699–4707.
Guo, L., W. Diao, C. Gao, G. Hu, Q. Ding, C. Ye, X. Chen, J. Liu, and L. Liu (2020) Engineering Escherichia coli lifespan for enhancing chemical production. Nat. Catal. 3: 307–318.
Liu, L., X. Duan, and J. Wu (2016) Modulating the direction of carbon flow in Escherichia coli to improve l-tryptophan production by inactivating the global regulator FruR. J. Biotechnol. 231: 141–148.
Mills, T. Y., N. R. Sandoval, and R. T. Gill (2009) Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol. Biofuels. 2: 26.
Fitzgerald, D. J., M. Stratford, M. J. Gasson, J. Ueckert, A. Bos, and A. Narbad (2004) Mode of antimicrobial action of vanillin against Escherichia coli, Lactobacillus plantarum and Listeria innocua. J. Appl. Microbiol. 97: 104–113.
Zaldivar, J., A. Martinez, and L. O. Ingram (1999) Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 65: 24–33.
Zaldivar, J., A. Martinez, and L. O. Ingram (2000) Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 68: 524–530.
Luo, H., P. Zheng, M. Bilal, F. Xie, Q. Zeng, C. Zhu, R. Yang, and Z. Wang (2020) Efficient bio-butanol production from lignocellulosic waste by elucidating the mechanisms of Clostridium acetobutylicum response to phenolic inhibitors. Sci. Total Environ. 710: 136399.
Zaldivar, J. and L. O. Ingram (1999) Effect of organic acids on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol. Bioeng. 66: 203–210.
Zhang, H. and G. Stephanopoulos (2013) Engineering E. coli for caffeic acid biosynthesis from renewable sugars. Appl. Microbiol. Biotechnol. 97: 3333–3341.
Luo, Z. W. and S. Y. Lee (2020) Metabolic engineering of Escherichia coli for the production of benzoic acid from glucose. Metab. Eng. 62: 298–311.
Lee, E. G., S. H. Yoon, A. Das, S. H. Lee, C. Li, J. Y. Kim, M. S. Choi, D. K. Oh, and S. W. Kim (2009) Directing vanillin production from ferulic acid by increased acetyl-CoA consumption in recombinant Escherichia coli. Biotechnol. Bioeng. 102: 200–208.
Cui, P., W. Zhong, Y. Qin, F. Tao, W. Wang, and J. Zhan (2020) Characterization of two new aromatic amino acid lyases from actinomycetes for highly efficient production of p-coumaric acid. Bioprocess Biosyst. Eng. 43: 1287–1298.
Barghini, P., D. Di Gioia, F. Fava, and M. Ruzzi (2007) Vanillin production using metabolically engineered Escherichia coli under non-growing conditions. Microb. Cell Fact. 6: 13.
Li, Y., J. Li, B. Qian, L. Cheng, S. Xu, and R. Wang (2018) De novo biosynthesis of p-coumaric acid in E. coli with a trans-cinnamic acid 4-hydroxylase from the Amaryllidaceae plant Lycoris aurea. Molecules. 23: 3185.
Lv, H., Y. Zhang, J. Shao, H. Liu, and Y. Wang (2021) Ferulic acid production by metabolically engineered Escherichia coli. Bioresour. Bioprocess. 8: 70.
Xu, Z., J. Kong, S. Zhang, T. Wang, and X. Liu (2020) Comparison of enzyme secretion and ferulic acid production by Escherichia coli expressing different Lactobacillus feruloyl esterases. Front. Microbiol. 11: 568716.
Pugh, S., R. McKenna, M. Osman, B. Thompson, and D. R. Nielsen (2014) Rational engineering of a novel pathway for producing the aromatic compounds p-hydroxybenzoate, protocatechuate, and catechol in Escherichia coli. Process Biochem. 49: 1843–1850.
Wu, J., G. Du, J. Zhou, and J. Chen (2013) Metabolic engineering of Escherichia coli for (2S)-pinocembrin production from glucose by a modular metabolic strategy. Metab. Eng. 16: 48–55.
Watts, K. T., P. C. Lee, and C. Schmidt-Dannert (2004) Exploring recombinant flavonoid biosynthesis in metabolically engineered Escherichia coli. Chembiochem. 5: 500–507.
Qi, W. W., T. Vannelli, S. Breinig, A. Ben-Bassat, A. A. Gatenby, S. L. Haynie, and F. S. Sariaslani (2007) Functional expression of prokaryotic and eukaryotic genes in Escherichia coli for conversion of glucose to p-hydroxystyrene. Metab. Eng. 9: 268–276.
Kim, B., H. Park, D. Na, and S. Y. Lee (2014) Metabolic engineering of Escherichia coli for the production of phenol from glucose. Biotechnol. J. 9: 621–629.
Yoon, S. H., C. Li, J. E. Kim, S. H. Lee, J. Y. Yoon, M. S. Choi, W. T. Seo, J. K. Yang, J. Y. Kim, and S. W. Kim (2005) Production of vanillin by metabolically engineered Escherichia coli. Biotechnol. Lett. 27: 1829–1832.
Chen, Z., X. Shen, J. Wang, J. Wang, Q. Yuan, and Y. Yan (2017) Rational engineering of p-hydroxybenzoate hydroxylase to enable efficient gallic acid synthesis via a novel artificial biosynthetic pathway. Biotechnol. Bioeng. 114: 2571–2580.
Kambourakis, S., K. M. Draths, and J. W. Frost (2000) Synthesis of gallic acid and pyrogallol from glucose: replacing natural product isolation with microbial catalysis. J. Am. Chem. Soc. 122: 9042–9043.
Verhoef, S., N. Wierckx, R. G. M. Westerhof, J. H. de Winde, and H. J. Ruijssenaars (2009) Bioproduction of p-hydroxystyrene from glucose by the solvent-tolerant bacterium Pseudomonas putida S12 in a two-phase water-decanol fermentation. Appl. Environ. Microbiol. 75: 931–936.
Wierckx, N. J. P., H. Ballerstedt, J. A. de Bont, and J. Wery (2005) Engineering of solvent-tolerant Pseudomonas putida S12 for bioproduction of phenol from glucose. Appl. Environ. Microbiol. 71: 8221–8227.
Nijkamp, K., R. G. Westerhof, H. Ballerstedt, J. A. de Bont, and J. Wery (2007) Optimization of the solvent-tolerant Pseudomonas putida S12 as host for the production of p-coumarate from glucose. Appl. Microbiol. Biotechnol. 74: 617–624.
Yamada, M., Y. Okada, T. Yoshida, and T. Nagasawa (2007) Biotransformation of isoeugenol to vanillin by Pseudomonas putida IE27 cells. Appl. Microbiol. Biotechnol. 73: 1025–1030.
Liu, H., X. Liu, H. Jiang, C. Liang, and Z. C. Zhang (2021) Enhanced lactic acid production from P(2)O(5)-pretreated biomass by domesticated Pediococcus pentosaceus without detoxification. Bioprocess Biosyst. Eng. 44: 2153–2166.
Wu, H., H. Wang, P. Wang, G. Zhao, H. Liu, L. Wang, X. Sun, and Z. Zheng (2021) Gradient radiation breeding and culture domestication of menaquinone producing strains. Bioprocess Biosyst. Eng. 44: 1373–1382.
Li, X.-J., R. Huang, C. Zhang, B. Luo, and D.-C. Gong (2014) Adaptation of a Saccharomyces Cerevisiae strain to lignocellulosic inhibitors by domestication. Sci. Technol. Food Ind. 7: 163–167.
Wang, M. (2018) Cultivation and domestication of activated sludge in village domestic sewage. Inn. Mong. Environ. Sci. 30: 58–59.
Yomano, L. P., S. W. York, and L. O. Ingram (1998) Isolation and characterization of ethanol-tolerant mutants of Escherichia coli KO11 for fuel ethanol production. J. Ind. Microbiol. Biotechnol. 20: 132–138.
Wang, L., B. Li, S.-P. Wang, Z.-Y. Xia, M. Gou, and Y.-Q. Tang (2021) Improving multiple stress-tolerance of a flocculating industrial Saccharomyces cerevisiae strain by random mutagenesis and hybridization. Process Biochem. 102: 275–285.
Nguyen, K. N., Y. Kim, S. Maibunkaew, J. Park, M. T. Nguyen, D.-B. Oh, and O. Kwon (2021) Enhanced production of 1-deoxynojirimycin in Bacillus subtilis subsp. inaquosorum by random mutagenesis and culture optimization. Biotechnol. Bioprocess Eng. 26: 265–276.
Gonzalez-Perez, D., J. Ratcliffe, S. K. Tan, M. Wong, Y. P. Yee, N. Nyabadza, J. H. Xu, T. S. Wong, and K. L. Tee (2021) Random and combinatorial mutagenesis for improved total production of secretory target protein in Escherichia coli. Sci. Rep. 11: 5290.
Yoon, S. H., E. G. Lee, A. Das, S. H. Lee, C. Li, H. K. Ryu, M. S. Choi, W. T. Seo, and S. W. Kim (2007) Enhanced vanillin production from recombinant E. coli using NTG mutagenesis and adsorbent resin. Biotechnol. Prog. 23: 1143–1148.
Yi, X., L. Lin, J. Mei, and W. Wang (2021) Transporter proteins in Zymomonas mobilis contribute to the tolerance of lignocellulose-derived phenolic aldehyde inhibitors. Bioprocess Biosyst. Eng. 44: 1875–1882.
Mitra, A., S. Chatterjee, S. Kataki, R. P. Rastogi, and D. K. Gupta (2021) Bacterial tolerance strategies against lead toxicity and their relevance in bioremediation application. Environ. Sci. Pollut. Res. Int. 28: 14271–14284.
Randhawa, A., N. Pasari, T. Sinha, M. Gupta, A. M. Nair, O. A. Ogunyewo, S. Verma, P. K. Verma, and S. S. Yazdani (2021) Blocking drug efflux mechanisms facilitate genome engineering process in hypercellulolytic fungus, Penicillium funiculosum NCIM1228. Biotechnol. Biofuels. 14: 31.
Segura, A., L. Molina, S. Fillet, T. Krell, P. Bernal, J. Muñoz-Rojas, and J. L. Ramos (2012) Solvent tolerance in Gramnegative bacteria. Curr. Opin. Biotechnol. 23: 415–421.
Bafna, J. A., E. Sans-Serramitjana, S. Acosta-Gutiérrez, I. V. Bodrenko, D. Hörömpöli, A. Berscheid, H. Brötz-Oesterhelt, M. Winterhalter, and M. Ceccarelli (2020) Kanamycin uptake into Escherichia coli is facilitated by OmpF and OmpC porin channels located in the outer membrane. ACS Infect. Dis. 6: 1855–1865.
Chetri, S., M. Singha, D. Bhowmik, K. Nath, D. D. Chanda, A. Chakravarty, and A. Bhattacharjee (2019) Transcriptional response of OmpC and OmpF in Escherichia coli against differential gradient of carbapenem stress. BMC Res. Notes. 12: 138.
Begic, S. and E. A. Worobec (2006) Regulation of Serratia marcescens ompF and ompC porin genes in response to osmotic stress, salicylate, temperature and pH. Microbiology (Reading). 152: 485–491.
Zhang, D. F., H. Li, X. M. Lin, S. Y. Wang, and X. X. Peng (2011) Characterization of outer membrane proteins of Escherichia coli in response to phenol stress. Curr. Microbiol. 62: 777–783.
Goose, J. E. and M. S. P. Sansom (2013) Reduced lateral mobility of lipids and proteins in crowded membranes. PLoS Comput. Biol. 9: e1003033.
Guzmán-Flores, J. E., L. Steinemann-Hernández, L. E. González de la Vara, M. Gavilanes-Ruiz, T. Romeo, A. F. Alvarez, and D. Georgellis (2019) Proteomic analysis of Escherichia coli detergent-resistant membranes (DRM). PLoS One. 14: e0223794.
Yin, N., G. Zhu, Q. Luo, J. Liu, X. Chen, and L. Liu (2020) Engineering of membrane phospholipid component enhances salt stress tolerance in Saccharomyces cerevisiae. Biotechnol. Bioeng. 117: 710–720.
Liu, Y., Q. Liu, A. Krivoruchko, S. Khoomrung, and J. Nielsen (2020) Engineering yeast phospholipid metabolism for de novo oleoylethanolamide production. Nat. Chem. Biol. 16: 197–205.
Keweloh, H., R. Diefenbach, and H. J. Rehm (1991) Increase of phenol tolerance of Escherichia coli by alterations of the fatty acid composition of the membrane lipids. Arch. Microbiol. 157: 49–53.
Satoh, S., M. Ozaki, S. Matsumoto, T. Nabatame, M. Kaku, T. Shudo, M. Asayama, and S. Chohnan (2020) Enhancement of fatty acid biosynthesis by exogenous acetyl-CoA carboxylase and pantothenate kinase in Escherichia coli. Biotechnol. Lett. 42: 2595–2605.
Cao, Y., J. Yang, M. Xian, X. Xu, and W. Liu (2010) Increasing unsaturated fatty acid contents in Escherichia coli by coexpression of three different genes. Appl. Microbiol. Biotechnol. 87: 271–280.
Liu, P., A. Chernyshov, T. Najdi, Y. Fu, J. Dickerson, S. Sandmeyer, and L. Jarboe (2013) Membrane stress caused by octanoic acid in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 97: 3239–3251.
Tan, Z., J. M. Yoon, D. R. Nielsen, J. V. Shanks, and L. R. Jarboe (2016) Membrane engineering via trans unsaturated fatty acids production improves Escherichia coli robustness and production of biorenewables. Metab. Eng. 35: 105–113.
Tan, Z., P. Khakbaz, Y. Chen, J. Lombardo, J. M. Yoon, J. V. Shanks, J. B. Klauda, and L. R. Jarboe (2017) Engineering Escherichia coli membrane phospholipid head distribution improves tolerance and production of biorenewables. Metab. Eng. 44: 1–12.
Jones, H. E., I. B. Holland, A. Jacq, T. Wall, and A. K. Campbell (2003) Escherichia coli lacking the AcrAB multidrug efflux pump also lacks nonproteinaceous, PHB-polyphosphate Ca2+ channels in the membrane. Biochim. Biophys. Acta. 1612: 90–97.
Rosenberg, E. Y., D. Ma, and H. Nikaido (2000) AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182: 1754–1756.
Licandro-Seraut, H., C. Roussel, G. Perpetuini, P. Gervais, and J. F. Cavin (2013) Sensitivity to vinyl phenol derivatives produced by phenolic acid decarboxylase activity in Escherichia coli and several food-borne Gram-negative species. Appl. Microbiol. Biotechnol. 97: 7853–7864.
Alekshun, M. N. and S. B. Levy (1999) Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181: 4669–4672.
Hächler, H., S. P. Cohen, and S. B. Levy (1991) marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 173: 5532–5538.
Van Dyk, T. K., L. J. Templeton, K. A. Cantera, P. L. Sharpe, and F. S. Sariaslani (2004) Characterization of the Escherichia coli AaeAB efflux pump: a metabolic relief valve? J. Bacteriol. 186: 7196–7204.
Kurgan, G., L. A. Panyon, Y. Rodriguez-Sanchez, E. Pacheco, L. M. Nieves, R. Mann, D. R. Nielsen, and X. Wang (2019) Bioprospecting of native efflux pumps to enhance furfural tolerance in ethanologenic Escherichia coli. Appl. Environ. Microbiol. 85: e02985–18.
Wang, S., X. Sun, and Q. Yuan (2018) Strategies for enhancing microbial tolerance to inhibitors for biofuel production: a review. Bioresour. Technol. 258: 302–309.
Turner, W. J. and M. J. Dunlop (2015) Trade-offs in improving biofuel tolerance using combinations of efflux pumps. ACS Synth. Biol. 4: 1056–1063.
Lee, K. and S. S. Yoon (2017) Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J. Microbiol. Biotechnol. 27: 1053–1064.
Valentini, M. and A. Filloux (2016) Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291: 12547–12555.
Li, Y., P. Xiao, Y. Wang, and Y. Hao (2020) Mechanisms and control measures of mature biofilm resistance to antimicrobial agents in the clinical context. ACS Omega. 5: 22684–22690.
Singh, R., D. Paul, and R. K. Jain (2006) Biofilms: implications in bioremediation. Trends Microbiol. 14: 389–397.
Farhadian, M., D. Duchez, C. Vachelard, and C. Larroche (2008) Monoaromatics removal from polluted water through bioreactors-a review. Water Res. 42: 1325–1341.
Wu, Y., Y. Ding, Y. Cohen, and B. Cao (2015) Elevated level of the second messenger c-di-GMP in Comamonas testosteroni enhances biofilm formation and biofilm-based biodegradation of 3-chloroaniline. Appl. Microbiol. Biotechnol. 99: 1967–1976.
Jia, X., S. Zhang, J. Li, J. Xia, R. Yao, X. Zhao, B. Wu, F. Bai, and Y. Xiao (2020) Engineered bacterial biofloc formation enhancing phenol removal and cell tolerance. Appl. Microbiol. Biotechnol. 104: 1187–1199.
Li, Q., X. Q. Zhao, A. K. Chang, Q. M. Zhang, and F. W. Bai (2012) Ethanol-induced yeast flocculation directed by the promoter of TPS1 encoding trehalose-6-phosphate synthase 1 for efficient ethanol production. Metab. Eng. 14: 1–8.
Wackett, L. P. (2003) Pseudomonas putida—a versatile biocatalyst. Nat. Biotechnol. 21: 136–138.
Wierckx, N. J., H. Ballerstedt, J. A. de Bont, and J. Wery (2005) Engineering of solvent-tolerant Pseudomonas putida S12 for bioproduction of phenol from glucose. Appl. Environ. Microbiol. 71: 8221–8227.
Wery, J., D. I. Mendes da Silva, and J. A. de Bont (2000) A genetically modified solvent-tolerant bacterium for optimized production of a toxic fine chemical. Appl. Microbiol. Biotechnol. 54: 180–185.
Verhoef, S., H. J. Ruijssenaars, J. A. de Bont, and J. Wery (2007) Bioproduction of p-hydroxybenzoate from renewable feedstock by solvent-tolerant Pseudomonas putida S12. J. Biotechnol. 132: 49–56.
Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-Gonzalez, A. Rojas, W. Teran, and A. Segura (2002) Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56: 743–768.
Ramos, J. L., E. Duque, J. J. Rodríguez-Herva, P. Godoy, A. Haïdour, F. Reyes, and A. Fernández-Barrero (1997) Mechanisms for solvent tolerance in bacteria. J. Biol. Chem. 272: 3887–3890.
Zhang, X.-Y., T.-T. Li, Y.-R. Liu, S.-S. Geng, A.-L. Luo, M.-S. Jiang, X.-W. Liang, J.-H. Shang, K.-H. Lu, and X.-G. Yang (2021) Transcriptome analysis revealed differences in the microenvironment of spermatogonial stem cells in seminiferous tubules between pre-pubertal and adult buffaloes. Reprod. Domest. Anim. 56: 629–641.
Lv, H., M. Kim, S. Park, K. Baek, H. Oh, J. E. W. Polle, and E. S. Jin (2021) Comparative transcriptome analysis of short-term responses to salt and glycerol hyperosmotic stress in the green alga Dunaliella salina. Algal Res. 53: 102147.
Gu, P., J. Kang, F. Yang, Q. Wang, Q. Liang, and Q. Qi (2013) The improved L-tryptophan production in recombinant Escherichia coli by expressing the polyhydroxybutyrate synthesis pathway. Appl. Microbiol. Biotechnol. 97: 4121–4127.
Dong, H., W. Zhang, Q. Xuan, Y. Zhou, S. Zhou, J. Huang, and P. Wang (2021) Binding peptide-guided immobilization of lipases with significantly improved catalytic performance using Escherichia coli BL21(DE3) biofilms as a platform. ACS Appl. Mater. Interfaces. 13: 6168–6179.
Müller, C., L. Petruschka, H. Cuypers, G. Burchhardt, and H. Herrmann (1996) Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J. Bacteriol. 178: 2030–2036.
Yang, J., T. Gao, Y. Zhang, S. Wang, H. Li, S. Li, and S. Wang (2019) Degradation of the phenolic β-ether lignin model dimer and dyes by dye-decolorizing peroxidase from Bacillus amyloliquefaciens. Biotechnol. Lett. 41: 1015–1021.
Acknowledgements
This work is supported by the National Natural Science Foundation of China (21808075), and the Natural Science Foundation of Jiangsu Province (BK20170459).
Author information
Authors and Affiliations
Contributions
LL conceived and designed research. XM, LW, and ST collected data. YZ, HL, and XD analyzed data. LL and MB wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
This article does not contain any studies with human participants or animals performed by any authors.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, L., Ma, X., Bilal, M. et al. Mechanistic Insight into Phenolic Compounds Toxicity and State-of-the-art Strategies for Enhancing the Tolerance of Escherichia coli to Phenolic Compounds. Biotechnol Bioproc E 27, 533–542 (2022). https://doi.org/10.1007/s12257-022-0019-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-022-0019-7