Abstract
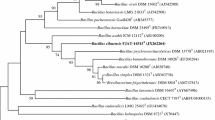
A Gram-positive, rod-shaped, motile, endospore-forming bacterial strain, designated NB22T, was isolated from soil of a lettuce field in Kyonggi province, South Korea, and was characterized by using a polyphasic taxonomic approach. This novel isolate grew optimally at 30–37°C and pH 8–9. It grew in the presence of 0–4% NaCl (optimum, 1–2%). Comparative 16S rRNA gene sequence analysis showed that strain NB22T was closely related to members of the genus Bacillus and fell within a coherent cluster comprising B. siralis 171544T (98.1%) and B. korlensis ZLC-26T (97.3%). The levels of 16S rRNA gene sequence similarity with respect to other Bacillus species with validly published names were less than 96.4%. Strain NB22T had a genomic DNA G+C content of 36.3 mol% and the predominant respiratory quinone was MK-7. The peptidoglycan contained meso-diaminopimelic acid. The major cellular fatty acids were iso-C15:0, anteiso-C15:0, C14:0, and C16:0. These chemotaxonomic results supported the affiliation of strain NB22T to the genus Bacillus, and the low DNA-DNA relatedness values and distinguishing phenotypic characteristics allowed genotypic and phenotypic differentiation of strain NB22T from recognized Bacillus species. On the basis of the evidence presented, strain NB22T is considered to represent a novel species of the genus Bacillus, for which the name Bacillus kyonggiensis sp. nov. is proposed. The type strain is NB22T (=KEMB 5401-267T =JCM 17569T).
Similar content being viewed by others
References
Baron, S. 1996. Chapter 15, Bacillus. In Medical Microbiology 4th edition. University of Texas Medical Branch at Galveston, Galveston, USA.
Chun, J., J.H. Lee, Y. Jung, M. Kim, S. Kim, B.K. Kim, and Y.W. Lim. 2007. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261.
Claus, D. and R.C.W. Berkeley. 1986. Genus Bacillus Cohn 1872. In Bergey’s Manual of Systematic Bacteriology, vol. 2, pp. 1105–1140. Williams and Wilkins, Baltimore, USA.
Doetsch, R.N. 1981. Determinative methods of light microscopy. In Manual of Methods for General Bacteriology, pp. 21–33. American Society for Microbiology, Washington, D.C., USA.
Euzéby, J.P. 2008. List of Prokaryotic Names with Standing in Nomenclature. http://www.bacterio.cict.fr/
Ezaki, T., Y. Hashimoto, and E. Yabuuchi. 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229.
Felsenstein, J. 1985. Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Hall, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hiraishi, A., Y. Ueda, J. Ishihara, and T. Mori. 1996. Comparative lipoquinone analysis of influent sewage and activated sludge by highperformance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 42, 457–469.
Kempf, M.J., F. Chen, R. Kern, and K. Venkateswaran. 2005. Recurrent isolation of hydrogen peroxide-resistant spores of Bacillus pumilus from a spacecraft assembly facility. Astrobiology 5, 391–405.
Kim, M.K., W.T. Im, H. Ohta, M. Lee, and S.T. Lee. 2005. Sphingopyxis granuli sp. nov., a β-glucosidase producing bacterium in the family Sphingomonadaceae in a-4 subclass of the Proteobacteria. J. Microbiol. 43, 152–157.
Kimura, M. 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press. Cambridge, UK.
Komagata, K. and K. Suzuki. 1987. Lipids and cell-wall analysis in bacterial systematics. Methods Microbiol. 19, 161–203.
Mesbah, M., U. Premachandran, and W.B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by highperformance liquid chromatography. Int. J. Syst. Bacteriol. 39, 159–167.
Minnikin, D.E., A.G. O’Donnell, M. Goodfellow, G. Anderson, M. Athalye, A. Schaal, and J.H. Parlett. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Sasser, M. 1990. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids, MIDI Technical Note 101. MIDI Inc., Newark, DE, USA.
Shida, O., H. Takagi, K. Kadowaki, L.K. Nakamura, and K. Komagata. 1997. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus. Int. J. Syst. Bacteriol. 47, 289–298.
Stackebrandt, E. and B.M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849.
Staneck, J.L. and G.D. Roberts. 1974. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 226–231.
Tamura, K., D. Peterson, N. Peterson, G. Stecher, M. Nei, and S. Kumar. 2011. MEGA 5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739.
Thompson, J.D., T.J. Gibson, F. Plewniak, F. Jeanmougin, and D.G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882.
Wayne, L.G., D.J. Brenner, R.R. Colwell, P.A.D. Grimont, O. Kandler, M.I. Krichevsky, L.H. Moore, and et al. 1987. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464.
Weisburg, W.G., S.M. Barns, D.A. Pelletier, and D.J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Wilson, K. 1987. Preparation of genomic DNA from bacteria. In Current Protocols in Molecular Biology, pp. 2.4.1–2.4.5. Green Publishing and Wiley-Interscience, New York, NY, USA.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dong, K., Lee, S. Bacillus kyonggiensis sp. nov., isolated from soil of a lettuce field. J Microbiol. 49, 776–781 (2011). https://doi.org/10.1007/s12275-011-1218-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-011-1218-7