Abstract
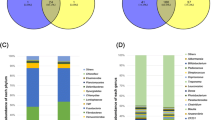
Gut microbial diversity and the core microbiota of the Jinhua pig, which is a traditional, slow-growing Chinese breed with a high body-fat content, were examined from a total of 105 fecal samples collected from 6 groups of pigs at 3 weaning ages that originated from 2 strains and were raised on 3 different pig farms. The bacterial community was analyzed following high-throughput pyrosequencing of 16S rRNA genes, and the fecal concentrations of short-chain fatty acids (SCFAs) were measured by gas chromatograph. Our results showed that Firmicutes and Bacteroidetes were the dominant phyla, and Lactobacillus, Streptococcus, Clostridium, SMB53, and Bifidobacterium were the most abundant genera. Fifteen predominant genera present in every Jinhua pig sample constituted a phylogenetic core microbiota and included the probiotics Lactobacillus and Bifidobacterium, and the SCFA-producing bacteria Clostridium, Prevotella, Bacteroides, Coprococcus, Roseburia, Ruminococcus, Blautia, and Butyricicoccus. Comparisons of the microbiota compositions and SCFA concentrations across the 6 groups of pigs demonstrated that genetic background and weaning age affected the structure of the gut microbiota more significantly than the farm. The relative abundance of the core genera in the pigs, including Lactobacillus, Clostridium, Prevotella, Bacteroides, Roseburia, Ruminococcus, Blautia, and Butyricicoccus varied dramatically in pigs among the 2 origins and 3 weaning ages, while Oscillospira, Megasphaera, Parabacteroides, and Corynebacterium differed among pigs from different farms. Interestingly, there was a more significant influence of strain and weaning age than of rearing farm on the SCFA concentrations. Therefore, strain and weaning age appear to be the more important factors shaping the intestinal microbiome of pigs.
Similar content being viewed by others
References
Backhed, F., Ley, R.E., Sonnenburg, J.L., Peterson, D.A., and Gordon, J.I. 2005. Host-bacterial mutualism in the human intestine. Science 307, 1915–1920.
Bell, D.S. 2015. Changes seen in gut bacteria content and distribution with obesity: causation or association? Postgrad. Med. 127, 863–868.
Bian, G.R., Ma, S.Q., Zhu, Z.G., Su, Y., Zoetendal, E.G., Mackie, R., Liu, J.H., Mu, C.L., Huang, R.H., Smidt, H., and Zhu, W.Y. 2016. Age, introduction of solid feed and weaning are more important determinants of gut bacterial succession in piglets than breed and nursing mother as revealed by a reciprocal cross-fostering model. Environ. Microbiol. 18, 1566–1577.
Bik, E.M. 2009. Composition and function of the human-associated microbiota. Nutr. Rev. 67 Suppl 2, S164–S171.
Blaut, M. 2015. Gut microbiota and energy balance: role in obesity. Proc. Nutr. Soc. 74, 227–234.
Brodziak, F., Meharg, C., Blaut, M., and Loh, G. 2013. Differences in mucosal gene expression in the colon of two inbred mouse strains after colonization with commensal gut bacteria. PLoS One 8, e72317.
Campbell, J.H., Foster, C.M., Vishnivetskaya, T., Campbell, A.G., Yang, Z.K., Wymore, A., Palumbo, A.V., Chesler, E.J., and Podar, M. 2012. Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 6, 2033–2044.
Franklin, M., Mathew, A., Vickers, J., and Clift, R. 2002. Characterization of microbial populations and volatile fatty acid concentrations in the jejunum, ileum, and cecum of pigs weaned at 17 vs 24 days of age. J. Anim. Sci. 80, 2904–2910.
Gibson, G.R. and Roberfroid, M.B. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125, 1401–1412.
Guo, J., Shan, T., Wu, T., Zhu, L.N., Ren, Y., An, S., and Wang, Y. 2011. Comparisons of different muscle metabolic enzymes and muscle fiber types in Jinhua and Landrace pigs. J. Anim. Sci. 89, 185–191.
Hildebrand, F., Nguyen, T.L., Brinkman, B., Yunta, R.G., Cauwe, B., Vandenabeele, P., Liston, A., and Raes, J. 2013. Inflammationassociated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 14, R4.
Hu, Y., Dun, Y., Li, S., Zhang, D., Peng, N., Zhao, S., and Liang, Y. 2015. Dietary Enterococcus faecalis LAB31 improves growth performance, reduces diarrhea, and increases fecal Lactobacillus number of weaned piglets. PLoS One 10, e0116635.
Isaacson, R. and Kim, H.B. 2012. The intestinal microbiome of the pig. Anim. Health Res. Rev. 13, 100–109.
Kim, H.B., Borewicz, K., White, B.A., Singer, R.S., Sreevatsan, S., Tu, Z.J., and Isaacson, R.E. 2011. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet. Microbiol. 153, 124–133.
Kim, H.B., Borewicz, K., White, B.A., Singer, R.S., Sreevatsan, S., Tu, Z.J., and Isaacson, R.E. 2012. Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin. Proc. Natl. Acad. Sci. USA 109, 15485–15490.
Kim, H.B. and Isaacson, R.E. 2015. The pig gut microbial diversity: Understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet. Microbiol. 177, 242–251.
Koenig, J.E., Spor, A., Scalfone, N., Fricker, A.D., Stombaugh, J., Knight, R., Angenent, L.T., and Ley, R.E. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 108 Suppl 1, 4578–4585.
Konstantinov, S.R. 2005. Lactobacilli in the porcine intestine: From composition to functionality. PhD Thesis Wageningen University, Wageningen, the Netherlands.
Konstantinov, S.R., Awati, A.A., Williams, B.A., Miller, B.G., Jones, P., Stokes, C.R., Akkermans, A.D., Smidt, H., and De Vos, W.M. 2006. Post-natal development of the porcine microbiota composition and activities. Environ. Microbiol. 8, 1191–1199.
Lamendella, R., Domingo, J.W., Ghosh, S., Martinson, J., and Oerther, D.B. 2011. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 11, 103.
Larsen, N., Vogensen, F.K., van den Berg, F.W., Nielsen, D.S., Andreasen, A.S., Pedersen, B.K., Al-Soud, W.A., Sorensen, S.J., Hansen, L.H., and Jakobsen, M. 2010. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5, e9085.
Ley, R.E., Backhed, F., Turnbaugh, P., Lozupone, C.A., Knight, R.D., and Gordon, J.I. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102, 11070–11075.
Ley, R.E., Peterson, D.A., and Gordon, J.I. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848.
Li, M., Bauer, L.L., Chen, X., Wang, M., Kuhlenschmidt, T.B., Kuhlenschmidt, M.S., Fahey, G.C., and Donovan, S.M. 2012. Microbial composition and in vitro fermentation patterns of human milk oligosaccharides and prebiotics differ between formula-fed and sow-reared piglets. J. Nutr. 142, 681–689.
Lu, X.M., Lu, P.Z., and Zhang, H. 2014. Bacterial communities in manures of piglets and adult pigs bred with different feeds revealed by 16S rDNA 454 pyrosequencing. Appl. Microbiol. Biotechnol. 98, 2657–2665.
Magoc, T. and Salzberg, S.L. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963.
Mathur, R. and Barlow, G.M. 2015. Obesity and the microbiome. Expert Rev. Gastroenterol. Hepatol. 9, 1087–1099.
Miao, Z.G., Wang, L.J., Xu, Z.R., Huang, J.F., and Wang, Y.R. 2009. Developmental changes of carcass composition, meat quality and organs in the Jinhua pig and Landrace. Animal 3, 468–473.
Pajarillo, E.A., Chae, J.P., Balolong, M.P., Kim, H.B., Seo, K.S., and Kang, D.K. 2014. Pyrosequencing-based analysis of fecal microbial communities in three purebred pig lines. J. Microbiol. 52, 646–651.
Pajarillo, E.A., Chae, J.P., Kim, H.B., Kim, I.H., and Kang, D.K. 2015a. Barcoded pyrosequencing-based metagenomic analysis of the faecal microbiome of three purebred pig lines after cohabitation. Appl. Microbiol. Biotechnol. 99, 5647–5656.
Pajarillo, E.A.B., Chae, J.P., Balolong, M.P., Kim, H.B., Seo, K.S., and Kang, D.K. 2015b. Characterization of the fecal microbial communities of Duroc pigs using 16S rRNA gene pyrosequencing. Asian-Australas. J. Anim. Sci. 28, 584–591.
Poroyko, V., White, J.R., Wang, M., Donovan, S., Alverdy, J., Liu, D.C., and Morowitz, M.J. 2010. Gut microbial gene expression in mother-fed and formula-fed piglets. PLoS One 5, e12459.
Rehman, A., Sina, C., Gavrilova, O., Hasler, R., Ott, S., Baines, J.F., Schreiber, S., and Rosenstiel, P. 2011. Nod2 is essential for temporal development of intestinal microbial communities. Gut 60, 1354–1362.
Riva, A., Borgo, F., Lassandro, C., Verduci, E., Morace, G., Borghi, E., and Berry, D. 2017. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ. Microbiol. 19, 95–105.
Schwiertz, A., Taras, D., Schafer, K., Beijer, S., Bos, N.A., Donus, C., and Hardt, P.D. 2010. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 18, 190–195.
Shanks, O.C., Kelty, C.A., Archibeque, S., Jenkins, M., Newton, R.J., McLellan, S.L., Huse, S.M., and Sogin, M.L. 2011. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl. Environ. Microbiol. 77, 2992–3001.
Shepherd, M.L., Swecker, W.S., Jensen, R.V., and Ponder, M.A. 2012. Characterization of the fecal bacteria communities of forage-fed horses by pyrosequencing of 16S rRNA V4 gene amplicons. FEMS Microbiol. Lett. 326, 62–68.
Spor, A., Koren, O., and Ley, R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279–290.
Sutherland, M.A., Niekamp, S.R., Rodriguez-Zas, S.L., and Salak-Johnson, J.L. 2006. Impacts of chronic stress and social status on various physiological and performance measures in pigs of different breeds. J. Anim. Sci. 84, 588–596.
Turnbaugh, P.J., Hamady, M., Yatsunenko, T., Cantarel, B.L., Duncan, A., Ley, R.E., Sogin, M.L., Jones, W.J., Roe, B.A., Affourtit, J.P., et al. 2009. A core gut microbiome in obese and lean twins. Nature 457, 480–484.
Wang, Q., Garrity, G.M., Tiedje, J.M., and Cole, J.R. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267.
Wu, T., Zhang, Z., Yuan, Z., Lo, L.J., Chen, J., Wang, Y., and Peng, J. 2013. Distinctive genes determine different intramuscular fat and muscle fiber ratios of the longissimus dorsi muscles in Jinhua and landrace pigs. PLoS One 8, e53181.
Xu, J. and Gordon, J.I. 2003. Honor thy symbionts. Proc. Natl. Acad. Sci.USA 100, 10452–10459.
Zhang, J., Guo, Z., Lim, A.A.Q., Zheng, Y., Koh, E.Y., Ho, D., Qiao, J., Huo, D., Hou, Q., and Huang, W. 2014. Mongolians core gut microbiota and its correlation with seasonal dietary changes. Sci. Rep. 4, 5001.
Zhou, J., Bruns, M.A., and Tiedje, J.M. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62, 316–322.
Zou, F., Zeng, D., Wen, B., Sun, H., Zhou, Y., Yang, M., Peng, Z., Xu, S., Wang, H., Fu, X., et al. 2016. Illumina Miseq platform analysis caecum bacterial communities of rex rabbits fed with different antibiotics. AMB Express 6, 100.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yang, H., Xiao, Y., Wang, J. et al. Core gut microbiota in Jinhua pigs and its correlation with strain, farm and weaning age. J Microbiol. 56, 346–355 (2018). https://doi.org/10.1007/s12275-018-7486-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-018-7486-8