Abstract
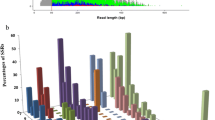
Microsatellite markers in small cardamom (Elettaria cardamomum Maton) were developed using the selective hybridization enrichment method. A total of 140 microsatellite repeats were identified from 270 clones. Primers were designed for 58 microsatellites and 44 primer pairs amplified products of expected size in cardamom. These markers were used for studying the diversity of 20 important small cardamom genotypes, and six markers were found to be polymorphic. The number of alleles ranged from 2 to 7 with an average of 3.6 per locus. Polymorphic information content values ranged from 0.14 to 0.38 based on dominant scoring. The two markers ECM 47a and ECMG 28 generated specific banding patterns for the genotypes MCC7 (Pink tiller) and APG434 (MA18) respectively. Dendrogram illustrated the genetic similarity between different genotypes of Kerala and Karnataka regions. It differentiated the closely related genotypes and released varieties into separate groups. Principal coordinate analysis revealed PV1 and ICRI 1 as the most divergent genotypes. The study demonstrated that these markers are informative and can be further utilized for generating reliable molecular data for assisting the crop improvement of small cardamom. Cross generic transferability (71.4 %) of the developed primers proved that they are useful for phylogenetic studies in the family Zingiberaceae. This is the first report of de novo isolation, characterisation and utilization of microsatellite markers for the genetic diversity analysis of small cardamom.



Similar content being viewed by others
References
Anu C, Ritto P, Prasath D, Deepesh PV, Nirmal Babu K, Parthasarathy VA (2015) Transferability of ginger, turmeric and large cardamom SSR primers to small cardamom (Elettaraia cardamomum Maton). J Trop Agric 53(2):107–115
Bassam JB, Gresshoff MP (2007) Silver staining DNA in Polyacrylamide gels. Nat Protoc 2(11):2649–2654
Blair MW, Panaud O, McCouch SR (1999) Inter-simple sequence repeat (lSSR) amplification for analysis of microsatellite motif frequency and fingerprinting in rice (Oryza sativa L). Theor Appl Genet 98:780–792
Bory S, Da Silva D, Risterucci AM, Grisoni M, Besse P, Duval MF (2008) Development of microsatellite markers in cultivated Vanilla: polymorphism and transferability to other Vanilla species. Sci Hortic 115:420–425
Das M, Banerjee S, Dhariwal R, Vyas S, Mir RR, Topdar N, Kundu A, Khurana JP, Tyagi AK, Sarkar D, Sinha MK, Balyan HS, Gupta PK (2012) Development of SSR markers and construction of a linkage map in jute. J Genet 91(1):21–31
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bul 19:11–15
Fan L, Zhang MY, Liu QZ, Li LT, Song Y, Wang LF, Zhang SL, Wu J (2013) Transferability of newly developed pear SSR markers to other Rosaceae species. Plant Mol Biol Rep 31:1271–1282
Fischer CEC (1956) Flora of the presidency of Madras. Part VIII. Ulmaceae to Xypridaceae. Government of India
Garris AJ, Tai TH, Cobum J, Kresovich S, McCouch S (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169:1631–1638
Gasic K, Han Y, Kertbundit S, Shulaev V, Iezzoni AF, Stover EW, Richard L, Michael B, Wisniewski E, Korban SS (2009) Characteristics and transferability of new apple EST-derived SSRs to other Rosaceae species. Mol Breed 23:397–411
Glenn TC, Schable NA (2005) Isolating microsatellite DNA loci. In: Methods enzymol 395:202–222. In: Zimmer EA, Roalson EH, (ed) Molecular evolution: producing the biochemical data, part b. Academic press, San Diego
Gregory PJ (1936) The floral morphology and cytology of Elettaria cardamomum Maton. J Linn Soc 50:362–391
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113:163–185
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl Acids Symp Se 41:95–98
Honig JA, Bonos SA, Meyer WA (2010) Isolation and characterization of 88 polymorphic microsatellite markers in kentucky bluegrass (Poa pratensis L.). HortScience 45(11):1759–1763
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–887
Joy N, Prasanth VP, Soniya EV (2011) Microsatellite based analysis of genetic diversity of popular black pepper genotypes in South India. Genetica 139(8):1033–1043
Katti MV, Ranjekar PK, Gupta VS (2001) Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol 18:1161–1167
Kress WJ, Linda MP, Kyle JW (2002) The phylogeny and a new classification of the gingers (Zingiberaceae): evidence from molecular data. Am J Bot 89(11):1682–1696
Kress WJ, Liu AZ, Newman M, Li QJ (2005) The molecular phylogeny of Alpinia (Zingiberaceae): a complex and polyphyletic genus of gingers. Am J Bot 92(1):167–178
Lee SY, Fai WK, Zakaria M, Ibrahim H, Othman RY, Gwag JG et al (2007) Characterization of polymorphic microsatellite markers, isolated from ginger (Zingiber officinale Rosc.). Mol Ecol Notes 7:1009–1011
Madhusoodanan KJ, Pradipkumar K, Ravindran PN (2002) Botany, crop improvement and biotechnology of cardamom. In: Ravindran PN, Madhusoodanan KJ (eds) Cardamom—the genus Elettaria. Taylor & Francis Inc, London, pp 11–68
Martins WS, Lucas DCS, Neves KFS, Bertioli DJ (2009) WebSat—A web software for microsatellite marker development. Bioinformation 3(6):282–283
Menezes IC, Cidade FW, Souza AP, Sampaio IC (2009) Isolation and characterization of microsatellite loci in the black pepper Piper nigrum L. (piperaceae). Conserv Genet Resour 1:209–212
Mir JI, Ahmed N, Khan MH, Mokhdomi TA, Wani SH, Bukhari S, Amin A, Qadri RA (2015) Molecular characterization of saffron-potential candidates for crop improvement. Not Sci Biol 7(1):81–89
Mun JH, Kim DJ, Choi HK, Gish J, Debelle F, Mudge J, Denny R, Endre G, Saurat O, Dudez AM, Kiss GB, Foe B, Young ND, Cook DR (2006) Distribution of microsatellites in the genome of Medicago truncatula: a resource of genetic markers that integrate genetic and physical maps. Genetics 172(4):2541–2555
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nirmal Babu K, Jayakumar VN, Minoo D, Venugopal MN, Sudarshan MR, Radhakrishnan VV, Backiyarani S, Narayanaswami MM, Peter KV, Parthasarathy VA (2012) Genetic diversity and phylogenetic relationships among small cardamom (Elettaria cardamomum Maton.) cultivars and related genera using DNA markers. Int J Innov Hortic 1(1):47–56
Park JY, Lee JK, Kim NS (2009) Simple sequence repeat polymorphisms (SSRPs) for evaluation of molecular diversity and germplasm classification of minor crops. Molecules 14:4546–4569
Prasath D, Venugopal MN (2004) Genetic diversity and conservation of cardamom (Elettaria cardamomum Maton) in India. Plant Genet Resour Newsl 138:55–60
Priyamedha Singh BK, Karur G, Sangha MK, Banga SS (2012) RAPD, ISSR and SSR based integrated linkage map from an F2 hybrid population of resynthesized and natural brassica carinata. Natl Acad Sci Lett 35(4):303–308
Radhakrishnan VV, Mohanan KV (2005) Molecular characterization of some elite genotypes of cardamom. Indian J Genet Plant Breed 65(3):227–228
Rana MK, Bhat KV (2004) A comparison of AFLP and RAPD markers for genetic diversity and cultivar identification in cotton. J Plant Biochem Biotechnol 13:19–24
Rohlf FJ (1998) NTSYSpc. Numerical taxonomy and multivariate analysis system. Version 2.02i. Applied Biostatistics, Inc, New York
Roldan-Ruiz I, Dendauw JE, Van bockstaele E, Depicker A, Loose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125–126
Rungis D, Llewellyn D, Dennis ES, Lyon BR (2005) Simple sequence repeat (SSR) markers reveal low levels of polymorphism between cotton (Gossypium hirsutum L.) cultivars. Aust J Agric Res 56(3):301–307
Sastri BN (1952) The wealth of India-Raw Materials, D-E, pp. 150–160
Sigrist MS, Pinheiro JB, Azevedo-Filho JA, Colombo CA (2009) Development and characterization of microsatellite markers for turmeric (Curcuma longa L.). Plant Breed 129:570–573
Siju S, Dhanya K, Syamkumar S, Sheeja TE, Sasikumar B, Bhatt AI, Parthasarathy VA (2010) Development, characterization and utilization of genomic microsatellite markers in turmeric (Curcuma longa L.). Biochem Syst Ecol 38:641–646
Tehrani MS, Mardi M, Saeidi H, Gharehyazi B, Assadi M (2008) Transferability of genomic and EST-microsatellites from Festuca arundinacea Schreb. to Lolium persicum Boiss. and Hohen. ex Boiss. Int J of Bot 4:476–480
Thiel T, Michalek W, Varshney RK, Garner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR markers in barley (Hordeum vulgare L.). Theor Appl Genet 106(3):411–422
Toth G, Gaspari Z, Jurka J (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10:967–981
Varshney RK, Kumar A, Balyan HS, Roy JK, Prasad M, Gupta PK (2000) Characterization of microsatellites and development of chromosome specific STMS markers in bread wheat. Plant Mol Biol Rep 18:5–16
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23(1):48–55
Venkateswarlu M, Urs SR, Nath BS, Shashidhar HE, Maheswaran M, Veeraiah TM, Sabitha MG (2006) A first genetic linkage map of mulberry (Morus spp.) using RAPD, ISSR, and SSR markers and pseudotestcross mapping strategy. Tree Genet Genomes 3(1):15–24
Wang ML, Barkley NA, Jenkins TM (2009) Microsatellite markers in plants and insects. Part I: applications of biotechnology. Genes, Genomes Genomics 3(1):54–67
Weir BS (1990) Genetic data analysis: methods for discrete population genetic data. Sinauer Associates Inc, Sunderland, MA
Yap IV, Nelson RJ (1996) WINBOOT: a program for performing bootstrap analysis for binary data to determine the confidence limits of UPGMA-based dendrograms. IRRI discussion series no. 14. International Rice Research Institute, Manila, Philippines
Acknowledgments
This work was supported by a research Grant from the Department of biotechnology (DBT), Government of India, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cyriac, A., Paul, R., Anupama, K. et al. Isolation and characterization of genomic microsatellite markers for small cardamom (Elettaria cardamomum Maton) for utility in genetic diversity analysis. Physiol Mol Biol Plants 22, 219–229 (2016). https://doi.org/10.1007/s12298-016-0355-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-016-0355-1