Abstract
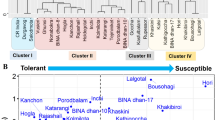
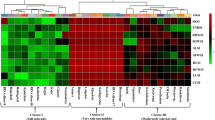
Salinity is a major abiotic stress that affects plant growth and development, especially in rice crop as it is a salt susceptible crop. Therefore, a wide range of rice genetic resources are screened in the germplasm banks to identify salt tolerant cultivars. The objective of this investigation was to develop effective indices for the classification of salt tolerant rice genotypes among Pathumthani 1, Khao Dawk Mali 105 (KDML 105), RD31, RD41, Suphanburi 1, RD43, RD49 and Riceberry. Rice seedlings were hydroponically grown with 10 dS m−1 NaCl treatment or without NaCl treatment (to serve as control) (WP; 2 dS m−1). Standard evaluation system peaked at a score of 9 in Pathumthani 1 and KDML 105, after 21 days of salt treatment, leading to leaf chlorosis, leaf burns and plant death. Chlorophyll a, chlorophyll b and total carotenoids were maintained better in the salt-stressed leaves of rice cvs. Riceberry and RD43, as compared to other cultivars. Salt stress induced a remarkable increase in the free proline accumulation (by 8.38 folds) in cv. Riceberry. Overall growth performance in rice cv. Riceberry was retained, whereas it declined in other cultivars. After 21 days of NaCl treatment at a concentration of 10 dS m−1, eight rice cultivars were classified into 3 groups based on multivariate physio-morphological indices, Group I: salt-tolerant rice, including cv. Riceberry; Group II: moderately salt tolerant, consisting of RD31, RD41, Suphanburi 1, RD43 and RD49 cultivars; Group III: salt-sensitive cultivars, namely Pathumthani 1 and KDML 105.





Similar content being viewed by others
References
Al-Aghabary K, Zhu Z, Shi Q (2005) Influence of silicon supply on chlorophyll content, chlorophyll fluorescence, and antioxidative in tomato plants under salt stress. J Plant Nutr 27:2101–2115
Ali MN, Yeasmin L, Gantait S, Goswami R, Chakraborty S (2014) Screening of rice landraces for salinity tolerance at seedling stage through morphological and molecular markers. Physiol Mol Biol Plant 20:411–423
Banu MNA, Hoque MA, Watanabe-Sugimoto M, Matsuoka K, Nakamura Y, Shioishi Y, Murata Y (2009) Proline and glycinebetaine induce antioxidant defense gene expression and suppress cell death in cultured tobacco cells under salt stress. J Plant Physiol 166:146–156
Barua R, de Ocampo M, Egdane J, Ismail AM, Mondal S (2015) Phenotyping rice (Oryza sativa L.) genotypes for physiological traits associated with tolerance of salinity at seedling stage. Sci Agric 12:156–162
Basu S, Giri RK, Kumar S, Rajwanshi R, Dwivedi SK, Kumar G (2017) Comprehensive physiological analyses and reactive oxygen species profiling in drought tolerant rice genotypes under salinity stress. Physiol Mol Biol Plant 23:837–850
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Cha-um S, Vejchasarn P, Kirdmanee C (2007) An effective defensive response in Thai aromatic rice varities (Oryza sativa L. spp. indica) to salinity. J Crop Sci Biotechnol 10:257–264
Cha-um S, Ashraf M, Kirdmanee C (2010) Screening upland rice (Oryza sativa L. ssp. indica) genotypes for salt-tolerance using multivariate cluster analysis. Afr J Biotechnol 9:4731–4740
Chunthaburee S, Sanitchon J, Pattanagul W, Theerakulpisut P (2014) Alleviation of salt stress in seedlings of black glutinous rice by seed priming with spermidine and gibberellic acid. Not Bot Horti Agrobot Cluj-Napoca 42:405–413
Chunthaburee S, Dongsansuk A, Sanitchon J, Pattanagul W, Theerakulpisut P (2016) Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt tolerance at seedling stage. Saudi J Biol Sci 23:467–477
Daiponmak W, Theerakulpisut P, Thanonkao P, Vanavichit A, Prathepha P (2010) Changes of anthocyanin cyaniding-3-glucoside content and antioxidant activity in Thai rice varieties under salinity stress. Sci Asia 36:286–291
de Leon TB, Lincombe S, Gregorio G, Subudhi K (2015) Genetic variation in Southern USA rice genotypes for seedling salinity tolerance. Front Plant Sci 6:374
Eynard A, Lal R, Wiebe K (2005) Crop response in salt-affected soils. J Sust Agric 27:5–50
Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010) Food security: the challenge of feeding 9 billion people. Science 327:812–817
Gregorio GB, Senadhira D, Mendoza RD (1997) Screening rice for salinity tolerance. IRRI Discussion Series No. 22. International Rice Research Institute, Manila, Philippines
Hariadi YC, Nurhayati AY, Soeparjono S, Arif I (2014) Screening six varieties of rice (Oryza sativa) for salinity tolerance. Proc Environ Sci 28:78–87
Huque MA, Okuma E, Banu MNA, Nakamura Y, Shimoishi Y, Murata Y (2007) Exogenous proline mitigates the detrimental effects of salt stress more than exogenous betaine by increasing antioxidant enzyme activity. J Plant Physiol 164:553–561
Kanawapee N, Sanitchon J, Lontom W, Threerakulpisut P (2012) Evaluation of salt tolerance at the seedlings stage in rice genotypes by growth performance, ion accumulation, proline and chlorophyll content. Plant Soil 358:235–249
Kordrostami M, Rabiei B, Kumleh HH (2017) Biochemical, physiological and molecular evaluation of rice cultivars differing in salt tolerance at seedling stage. Physiol Biol Plant 23:529–544
Kranto S, Chankaew S, Monkham T, Theerakulpisut P, Sanichon J (2016) Evaluation for salt tolerance in rice using multiple screening methods. J Agric Technol 18:1921–1931
Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Method Enzymol 148:350–382
Mohammadi R, Mendioro MS, Diaz GQ, Gregorio GB, Singh RK (2014) Genetic analysis of salt tolerance at seedling and reproductive stages in rice (Oryza sativa). Plant Breed 133:548–559
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Omisun T, Sahoo S, Saha B, Panda SK (2018) Relative salinity tolerance of rice cultivars native to North East India: a physiological, biochemical and molecular perspective. Protoplasma 255:193–202
Polle A, Chen S (2015) On the salty side of life: molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ 38:1794–1816
Pongprayoon W, Cha-um S, Pichakum A, Kirdmanee C (2008) Proline profiles in aromatic cultivars photoautotrophically grown in responses to salt stress. Int J Bot 4:276–282
Qadir M, Quillérou E, Nangia V, Murtaza G, Singh M, Thomas RJ, Drechsel P, Noble AD (2014) Economics of salt-induced land degradation and restoration. Nat Resour Forum A U N Sust Dev J 38:282–295
Ravikiran RT, Krishnamurthy SL, Warraich AS, Sharma PC (2018) Diversity and haplotypes of rice genotypes for seedling stage salinity tolerance analyzed through morpho-physiological and SSR markers. Field Crop Res 220:10–18
Sakina A, Ahmed I, Shahzad A, Iqbal M, Asif M (2015) Genetic variation for salinity tolerance in Pakistani rice (Oryza sativa L.) germplasm. J Agron Crop Sci 202:25–36
Shabala SN, Shabala SI, Martynenko AI, Babourina O, Newman IA (1998) Salinity effect on bioelectric activity, growth, Na+ accumulation and chlorophyll fluorescence of maize leaves: a comparative survey and prospects for screening. Aust J Plant Physiol 25:609–616
Vajrabhaya M, Vajrabhaya T (1991) Somaclonal variation of salt tolerance in rice. In: Bajaj YPS (ed) Biotechnology in agriculture and forestry 14. Springer, Berlin, pp 368–382
Wanichananan P, Kirdmanee C, Vutiyano C (2003) Effect of salinity on biochemical and physiological characteristics in correlation to selection of salt-tolerance in aromatic rice (Oryza sativa L.). Sci Asia 29:333–339
Wankhade SD, Cornejo MJ, Aateu-Andres I, Sanz A (2013) Morpho-physiological variaations in response to NaCl stress during vegetative and reproductive development of rice. Acta Physiol Plant 35:323–333
Zeng L (2005) Exploration of relationships between physiological parameters and growth performance of rice (Oryza sativa L.) seedlings under salinity stress using multivariate analysis. Plant Soil 268:51–59
Zeng L, Shannon MC, Grieve CM (2002) Evaluation of salt tolerance in rice genotypes by multiple agronomic parameters. Euphytica 127:235–245
Zeng L, Poss JA, Wilson C, Draz ASE, Gregorio GB, Grieve CM (2003) Evaluation of salt tolerance in rice genotypes by physiological characters. Euphytica 129:281–292
Acknowledgements
The authors are grateful to Suphan Buri Rice Research Center (Rice Research Institute, Department of Agriculture, Ministry of Agriculture and Cooperative, Thailand) for providing rice seeds. We would like to thank Faculty of Science, Burapha University and National Center for Genetic Engineering and Biotechnology (BIOTEC) for funding support and lab facilities.
Author information
Authors and Affiliations
Contributions
WP conducted the research project, report to funding agency, analyzed the data and wrote a first draft of manuscript, CT performed the experiment layout, free proline assay and analysis, RT analyzed the cluster ranking, critical revision of the data and performed the experiment, and SC performed the secondary data analysis and finished the final version of manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Pongprayoon, W., Tisarum, R., Theerawittaya, C. et al. Evaluation and clustering on salt-tolerant ability in rice genotypes (Oryza sativa L. subsp. indica) using multivariate physiological indices. Physiol Mol Biol Plants 25, 473–483 (2019). https://doi.org/10.1007/s12298-018-00636-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-018-00636-2