Abstract
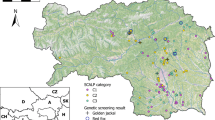
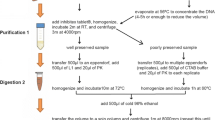
Identification of the species is a crucial step in many ecological studies. Sometimes, this could become a challenge, because of animal elusive behavior, low density population or for sympatric species leaving similar signs that are impossible to discriminate based only on their morphology. Here, we set up non-invasive molecular method to discern between the Italian hare (Lepus corsicanus) and the European hare (Lepus europaeus) using High Resolution Melting assay on fecal DNA, for the first time on these species. The Italian hare is endemic of the Central-Southern Italy and Sicily and it is classified vulnerable by the International Union for Conservation of Nature. Our procedure could be a useful tool to help conservation and management strategies, mainly in areas where the Italian hare and the European hare live in sympatry. The 9.5% of the peninsular range distribution of the European hare is took up by the range of the Italian hare. Our workflow allows sure species discrimination, rapidly and inexpensively (in one day at least 36 samples could be processed at costs of about 259 euros, including both DNA extractions and HRM run), also when large numbers of samples have to be processed. Moreover, our method could be widely applicable to other Lepus and/or mammalian species with similar concerns, by small adjustments to the protocol and its further validation, focusing on primes and corresponding HRM annealing temperature.



Similar content being viewed by others
References
Angelici F, Randi E, Riga F, Trocchi V (2008) Lepus corsicanus. The IUCN Red List of Threatened Species Version 20151 https://www.iucnredlistorg
Ball MC, Pither R, Manseau M, Clark J, Petersen SD, Kingston S, Morrill N, Wilson P (2007) Characterization of target nuclear DNA from faeces reduces technical issues associated with the assumptions of low-quality and quantity template. Conserv Genet 8:577–586
Banks SC, Piggott MP, Hansen BD, Robinson NA, Taylor AC (2002) Wombat coprogenetics enumerating a common wombat population by microsatellite analysis of faecal DNA. Aust J Zool 50:193–204
Berry O, Sarre SD (2007) Gel-free species identification using melt-curve analysis. Mol Ecol Notes 7:1–4
Brinkman TJ, Schwartz MK, Person DK, Pilgrim KL, Hundertmark KJ (2010) Effects of time and rainfall on PCR success using DNA extracted from deer fecal pellets. Conserv Genet 11:1547–1552
Buglione M (2017). Studio sulla nicchia ecologica di lepre italica (Lepus corsicanus). Ph Doctorate thesis. University of Naples Federico II.
Buglione M, Maselli V, Rippa D, de Filippo G, Trapanese M, Fulgione D (2018) A pilot study on the application of DNA metabarcoding for non-invasive diet analysis in the Italian hare. Mam Biol 88:31–42
Chen MT, Liang YJ, Kuo CC, Pei KJC (2016) Home ranges, movements and activity patterns of leopard cats (Prionailurus bengalensis) and threats to them in Taiwan. Mam Study 41:77–87
Dalén L, Götherström A, Angerbjörn A (2004) Identifying species from pieces of faeces. Conserv Genet 5:109–111
Davison A, Birks JD, Brookes RC, Braithwaite TC, Messenger JE (2002) On the origin of faeces: morphological versus molecular methods for surveying rare carnivores from their scats. J Zool 257:141–143
Deagle BE, Eveson JP, Jarman SN (2006) Quantification of damage in DNA recovered from highly degraded samples–a case study on DNA in faeces. Front Zool 3:11
De Barba M, Miquel C, Boyer F, Mercier C, Rioux D, Coissac E, Taberlet P (2014) DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet. Mol Ecol Resour 14:306–323
de Filippo G, Caliendo MF, Fulgione D, Fusco L, Troisi SR (2007) Status delle popolazioni di Lepus corsicanus nel territorio del Parco Nazionale del Cilento e Vallo di Diano de Filippo G. De Riso L, Riga F, Trocchi V and Troisi SR (a cura di)
Delibes-Mateos M, Rouco C, Villafuerte R (2009) Can adult and juvenile rabbits be differentiated by their pellet sizes? Acta Oecol 35:250–252
Drummond AJ (2011) Geneious v5 4 https://www.geneious.com
Edgar GJ, Barrett NS, Morton AJ (2004) Biases associated with the use of underwater visual census techniques to quantify the density and size-structure of fish populations. J Exp Mar Biol Ecol 308:269–290
Farrar JS, Wittwer CT (2017) High-resolution melting curve analysis for molecular diagnostics. In: Farrar JS, Wittwer CT (eds) molecular diagnostics. Academic Press, Cambridge, pp 79–102
Farrell LE, Roman J, Sunquist ME (2000) Dietary separation of sympatric carnivores identified by molecular analysis of scats. Mol Ecol 9:1583–1590
Flagstad O, Roed K, Stacy JE, Jakobsen KS (1999) Reliable noninvasive genotyping based on excremental PCR of nuclear DNA purified with a magnetic bead protocol. Mol Ecol 8:879–883
Fulgione D, Maselli V, Pavarese G, Rippa D, Rastogi RK (2009) Landscape fragmentation and habitat suitability in endangered Italian hare (Lepus corsicanus) and European hare (Lepus europaeus) populations European. J Wildl Res 55:385–396
Fusco L, Trosi SR, Accardo Y, Vaccaro L, Caliendo MF, de Filippo G (2007) Interspecific habitat selection between Italic and European Hares sympatric populations. In: V European Congress of Mammalogy pp 21–26
Garcia-Mazcorro JF, Castillo-Carranza SA, Guard B, Gomez-Vazquez JP, Dowd SE, Brigthsmith DJ (2017) Comprehensive molecular characterization of bacterial communities in feces of pet birds using 16S marker sequencing. Microb Ecol 73(1):224–235
Hansen MM, Jacobsen L (1999) Identification of mustelid species: otter (Lutra lutra), American mink (Mustela vison) and polecat (Mustela putorius), by analysis of DNA from faecal samples. J Zool 247:177–181
Hunter ME, Oyler-McCance SJ, Dorazio RM, Fike JA, Smith BJ, Hunter CT, Hart KM (2015) Environmental DNA (eDNA) sampling improves occurrence and detection estimates of invasive Burmese pythons. PLoS ONE 10:e0121655
Kasapidis P, Suchentrunk F, Magoulas A, Kotoulas G (2005) The shaping of mitochondrial DNA phylogeographic patterns of the brown hare (Lepus europaeus) under the combined influence of Late Pleistocene climatic fluctuations and anthropogenic translocations. Mol Phylogenet Evol 34:55–66
Kohn MH, Wayne RK (1997) Facts from feces revisited. Trends in Ecol Evol 12:223–227
Murphy MA, Waits LP, Kendall KC (2000) Quantitative evaluation of fecal drying methods for brown bear DNA analysis. Wildlife Soc B 28:951–957
Nechvatal JM, Ram JL, Basson MD, Namprachan P, Niec SR, Badsha KZ, Matherly LH, Majumdarh APN, Kato I (2008) Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiolo Methods 72:124–132
Oakley BB, Lillehoj HS, Kogut MH, Kim WK, Maurer JJ, Pedroso A, Cox NA (2014) The chicken gastrointestinal microbiome. FEMS Microbiol Lett 360:100–112
Palomares F, Godoy JA, Píriz A, O'Brien SJ, Johnson WE (2002) Fecal genetic analysis to determine the presence and distribution of elusive carnivores: design and feasibility for the Iberian lynx. Mol Ecol 11:2171–2182
Pan D, Yu Z (2014) Intestinal microbiome of poultry and its interaction with host and diet. Gut microbes 5:108–119
Pierpaoli M, Riga F, Trocchi V, Randi E (1999) Species distinction and evolutionary relationships of the Italian hare (Lepus corsicanus) as described by mitochondrial DNA sequencing. Mol Ecol 8:1805–1817
Pierpaoli M, Riga F, Trocchi V, Randi E (2003) Hare populations in Europe: intra and interspecific analysis of mtDNA variation. C R Biol 326:80–84
Pietri C, Alves PC, Melo-Ferreira J (2011) Hares in Corsica: high prevalence of Lepus corsicanus and hybridization with introduced L europaeus and L granatensis. Eur J Wildl Res 57:313–321
Pietri C (2015) Range and status of the Italian hare Lepus corsicanus in Corsica. Hystrix 26:2
Piggott MP (2005) Effect of sample age and season of collection on the reliability of microsatellite genotyping of fecal DNA. Wildl Res 31:485–493
Perry GH, Marioni JC, Melsted P, Gilad Y (2010) Genomic-scale capture and sequencing of endogenous DNA from feces. Mol Ecol 19:5332–5344
Ramón-Laca A, Gleeson D, Yockney I, Perry M, Nugent G, Forsyth DM (2014) Reliable discrimination of 10 ungulate species using high resolution melting analysis of fecal DNA. PLoS ONE 9:e92043
Randi E, Riga F (2019) Lepus corsicanus. The IUCN Red list of threatened species 2019: eT41305A2952954 https://dx.doi.org/10.2305/IUCNUK2019-2RLTST41305A2952954en
Reed GH, Kent JO, Wittwer CT (2007) High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 8(6):597–608
Reed JZ, Tollit DJ, Thompson PM, Amos W (1997) Molecular scatology: the use of molecular genetic analysis to assign species, sex and individual identity to seal faeces. Mol Eco 6:225–234
Reisser J, Proietti M, Kinas P, Sazima I (2008) Photographic identification of sea turtles: method description and validation, with an estimation of tag loss. Endang Species Res 5:73–82
Riga F, Trocchi V, Randi E, Toso S (2001) Morphometric differentiation between the Italian hare (Lepus corsicanus De Winton, 1898) and the European brown hare (Lepus europaeus Pallas, 1778). J Zool 253:241–252
Rouco C, Delibes-Mateos M, Moreno S (2009) Evidence against the use of fecal pellet size for age determination in European wild rabbits. Acta Oecol 35:668–670
Rouco C, Starkloff A, Delibes-Mateos M, Schubert M, Rödel HG (2012) Differentiation of animals from different age classes by means of pellet size: Assessment of a field method in European rabbits. Mammal Biol 77:451–454
Ruppert KM, Kline RJ, Rahman MS (2019) Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob Ecol Conserv 17:e00547
Santini A, Lucchini V, Fabbri E, Randi E (2007) Ageing and environmental factors affect PCR success in wolf (Canis lupus) excremental DNA samples. Mol Ecol Notes 7:955–961
Taberlet P, Coissac E, Hajibabaei M, Rieseberg LH (2012) Environmental DNA. Mol Ecol 21:1789–1793
Taberlet P, Waits LP, Luikart G (1999) Noninvasive genetic sampling: look before you leap. Trends Ecol Evol 14:323–327
Trocchi V, Riga F (2005). I lagomorfi in Italia. Linee guida per la conservazione e la gestione. Ministero delle Politiche Agricole e Forestali–Istituto Nazionale Fauna Selvatica. Documenti Tecnici : 1–128
Valentini A, Pompanon F, Taberlet P (2009) DNA barcoding for ecologists. Trends Ecol Evol 24:110–117
Vossen RH, Aten E, Roos A, den Dunnen JT (2009) High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat 30:860e866
Vynne C, Baker MR, Breuer ZK, Wasser SK (2012) Factors influencing degradation of DNA and hormones in maned wolf scat. Anim Conserv 15:184–194
Wasser SK, Houston CS, Koehler GM, Cadd GG, Fain SR (1997) Techniques for application of fecal DNA methods to field studies of Ursids. Mol Ecol 6:1091–1097
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ (2003) High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem 49:853–860
Acknowledgements
We thank the Cilento, Vallo di Diano e Alburni National Park (Salerno, Italy) for facilities. We are also grateful to Emeritus Dr. S.P. Goyal and one anonymous review for their helpful suggestions.
Author information
Authors and Affiliations
Contributions
Conceptualization: Maria Buglione and Domenico Fulgione; Methodology: Maria Buglione, Domenico Fulgione; Sample collection: Maria Buglione, Domenico Fulgione, Simona Petrelli, Tommaso Notomista and Gabriele de Filippo; Formal analysis: Maria Buglione and Domenico Fulgione; Investigation: Maria Buglione, Domenico Fulgione and Simona Petrelli; Resources: Domenico Fulgione, Maria Buglione, Simona Petrelli, Tommaso Notomista, Gabriele de Filippo and Romano Gregorio; Supervision: Maria Buglione and Domenico Fulgione; Writing – original draft: Maria Buglione and Domenico Fulgione; Writing – review & editing: Maria Buglione, Domenico Fulgione, Simona Petrelli, Tommaso Notomista and Gabriele de Filippo and Romano Gregorio.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Buglione, M., Petrelli, S., Notomista, T. et al. Who is who? High Resolution Melting analysis to discern between hare species using non-invasive sampling. Conservation Genet Resour 12, 727–732 (2020). https://doi.org/10.1007/s12686-020-01153-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-020-01153-9