Abstract
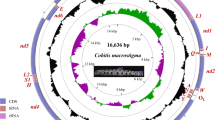
The suborder Charadrii (Aves: Charadriiformes), one of the most species-rich radiations within shorebirds, which contains good source for studies of ecology, behaviour and evolution. The resources of mitogenome have rapidly accumulated in recent years due to the advanced genomic sequencing, while suborder Charadrii’s mitogenome has not been well studied. The primary objective of this study was to determine the complete mitogenome sequence of Charadrius alexandrinus, and investigated the evolutionary relationship within Charadrii. The mitogenome of C. alexandrinus were generated by amplification of overlapping Polymerase Chain Reaction (PCR) fragments. In this study, we determined the complete mitogenome sequence of the Kentish Plover Charadrius alexandrinus, and comparative analysed 11 species to illustrate mitogenomes structure and investigated their evolutionary relationship within Charadrii. The Charadrii mitogenomes displayed moderate size variation, the mean size was 16,944 bp (SD = 182, n = 11), and most of the size variation due to mutations in the control region (CR). Nucleotide composition was consistently biased towards AT rich, and the A+T content also varies for each protein-coding genes. The variation in ATP8 and COIII was the highest and lowest respectively. The GC skew was always negative, with the ATP8 had higher value than other regions. The average uncorrected pairwise distances revealed heterogeneity of evolutionary rate for each gene, the COIII, COI and COII have slow evolutionary rate, whereas the gene of ATP8 has the relative fast rate. The highest value of Ks and Ka were ND1 and ATP8, and the ratios of Ka/Ks are lower than 0.27, indicating that they were under purifying selection. Phylogenomic analysis based on the complete mitochondrial genomes strongly supported the monophyly of the suborder Charadrii. This study improves our understanding of mitogenome structure and evolution, and providing further insights into phylogeny and taxonomy in Charadrii. In future, sequencing more mitogenomes from various taxonomic levels will significantly improve our understanding of phylogenetic relationships within Charadrii.




Similar content being viewed by others
References
Almalki M et al (2017) Morphological and genetic differentiation among kentish plover Charadrius alexandrinus populations in Macaronesia. Ardeola 64:3–16
Baker AJ, Pereira SL, Paton TA (2007) Phylogenetic relationships and divergence times of Charadriiformes genera: multigene evidence for the Cretaceous origin of at least 14 clades of shorebirds. Biol Lett 3:205–209
Baker AJ, Yatsenko Y, Tavares ES (2012) Eight independent nuclear genes support monophyly of the plovers: the role of mutational variance in gene trees. Mol Phylogenet Evol 65:631–641
Ericson PG, Envall I, Irestedt M, Norman JA (2003) Inter-familial relationships of the shorebirds (Aves: Charadriiformes) based on nuclear DNA sequence data. BMC Evol Biol 3:149–154
Fain MG, Houde P (2007) Multilocus perspectives on the monophyly and phylogeny of the order Charadriiformes (Aves). BMC Evol Biol 7:324
Friesen VL (2015) Speciation in seabirds: why are there so many species… and why aren’t there more? J Ornithol 156:1–13
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment program for Windows 95/98/NT. Nucleic acids symposium series no 41, pp 95–98
Hassanin A (2006) Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol 38:100–116
Hassanin A, Léger N, Deutsch J (2005) Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst Biol 54:277–298
Hu C, Zhang C, Lei S, Yi Z, Xie W, Zhang B, Chang Q (2017) The mitochondrial genome of pin-tailed snipe Gallinago stenura, and its implications for the phylogeny of Charadriiformes. PLoS ONE 12:e0175244
Küpper C et al (2012) High gene flow on a continental scale in the polyandrous Kentish plover Charadrius alexandrinus. Mol Ecol 21:5864–5879
Lanfear R, Calcott B, Ho SY, Guindon S (2012) Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29:1695–1701
Laslett D, Canbäck B (2008) ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24:172–175
Lavrov DV, Boore JL, Brown WM (2002) Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: duplication and nonrandom loss. Mol Biol Evol 19:163–169
Li X, Huang Y, Lei F (2014) Comparative mitochondrial genomics and phylogenetic relationships of the Crossoptilon species (Phasianidae, Galliformes). BMC Genom 16:1–12
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lobry JR (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol 13:660–665
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41:575–581
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Marshall HD, Baker AJ, Grant AR (2013) Complete mitochondrial genomes from four subspecies of common chaffinch (Fringilla coelebs): new inferences about mitochondrial rate heterogeneity, neutral theory, and phylogenetic relationships within the order Passeriformes. Gene 517:37–45
Mayr G (2011) The phylogeny of Charadriiform birds (shorebirds and allies): reassessing the conflict between morphology and molecules. Zool J Linn Soc 161:916–934
Mindell DP, Sorenson MD, Dimcheff DE (1998) An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol 15:1568–1571
Pacheco MA, Battistuzzi FU, Lentino M, Aguilar RF, Kumar S, Escalante AA (2011) Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders. Mol Biol Evol 28:1927–1942
Paton TA, Baker AJ (2006) Sequences from 14 mitochondrial genes provide a well-supported phylogeny of the Charadriiform birds congruent with the nuclear RAG-1 tree. Mol Phylogenet Evol 39:657–667
Paton TA, Baker AJ, Groth JG, Barrowclough GF (2003) RAG-1 sequences resolve phylogenetic relationships within Charadriiform birds. Mol Phylogenet Evol 29:268–278
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358
Que P, Chang Y, Eberhart-Phillips L, Liu Y, Székely T, Zhang Z (2015) Low nest survival of a breeding shorebird in Bohai Bay, China. J Ornithol 156:297–307
Ren Q et al (2014) The complete mitochondrial genome of the yellow-browed bunting, Emberiza chrysophrys (Passeriformes: Emberizidae), and phylogenetic relationships within the genus Emberiza. J Genet 93:699–707
Sambrook J, Russell DW (1989) Molecular cloning:a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Smith NA, Clarke JA (2015) Systematics and evolution of the Pan-Alcidae (Aves, Charadriiformes). J Avian Biol 46:125–140
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449
Yoon KB, Cho CU, Park YC (2015) The mitochondrial genome of the Saunders’s gull Chroicocephalus saundersi (Charadriiformes: Laridae) and a higher phylogeny of shorebirds (Charadriiformes). Gene 572:227–236
Acknowledgements
We wish to thank Peng Chen for kindly collecting samples, Yunhao Wu and Deyun Tai for data analysis, and Yi Zhang for the help with laboratory work. This research was supported by Grants from the Natural Science Research Program of Jiangsu Higher Education Institutions of China (Grant Nos. 16KJB180003, 16KJB180012); ‘12th Five-Year’ Planning Issue of Jiangsu Open University (The City Vocational College of Jiangsu) (Grant No. 15SEW-Y-018); Training program of innovation and entrepreneurship for undergraduates in Jiangsu, China (Grant No. 201714000015X); and Nature Science Fund of Jiangsu Province (Grant No. BK20161480).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, W., Zhang, C., Pan, T. et al. The mitochondrial genome of the Kentish Plover Charadrius alexandrinus (Charadriiformes: Charadriidae) and phylogenetic analysis of Charadrii. Genes Genom 40, 955–963 (2018). https://doi.org/10.1007/s13258-018-0703-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-018-0703-3