Abstract
MicroRNA (miRNA) deregulation is associated with various cancers. Among an expanding list of cancer-related miRNAs, deregulation of miR-125b has been well documented in many cancers including breast. Based on current knowledge, miR-125b is considered to be a tumor suppressor in breast cancers. While important messenger RNA (mRNA) targets have been defined for miR-125b, here, we aimed to further investigate direct/indirect consequences of miR-125b expression in breast cancer cells by using a transcriptome approach. Upon miR-125b expression, a total of 138 cancer-related genes were found to be differentially expressed in breast cancer cells. While only a few of these were predicted to be direct mRNA targets, majority of the gene expression changes were potentially downstream and indirect effects of miR-125b expression. Among these, activated leukocyte antigen molecule (ALCAM) mRNA and protein levels were found to be highly significantly increased upon miR-125b expression. Given the tumor suppressor role of miR-125b in our model system, upon silencing of ALCAM expression, cell proliferation rate re-increased in miR-125b-expressing cells. While ALCAM’s possible context-dependent roles are not clear in breast cancer, a diverse expression pattern of ALCAM mRNA was detected in a panel of breast cancer patient samples. Differentially expressed/regulated cancer-related genes upon miR-125b expression along with the significant increase of ALCAM are of future interest to understand how deregulated expression of miR-125b may have a tumor suppressor role in breast and other cancers.





Similar content being viewed by others
References
Bartel D. Micrornas: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Erson A, Petty E. MicroRNAs in development and disease. Clin Genet. 2008;74:296–306.
Erson AE, Petty EM. MiRNAs and cancer: new research developments and potential clinical applications. Cancer Biol Ther. 2009;8:2317–22.
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70.
Calin G, Croce C. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Iorio M, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707.
Bloomston M, Frankel W, Petrocca F, Volinia S, Alder H, Hagan J, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8.
Bousquet M, Quelen C, Rosati R, Mansat-De Mas V, La Starza R, Bastard C, et al. Myeloid cell differentiation arrest by mir-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–506.
Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–7.
Selcuklu SD, Yakicier MC, Erson AE. An investigation of microRNAs mapping to breast cancer related genomic gain and loss regions. Cancer Genet Cytogenet. 2009;189:15–23.
Mattie M, Benz C, Bowers J, Sensinger K, Wong L, Scott G, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24.
Scott G, Goga A, Bhaumik D, Berger C, Sullivan C, Benz C. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA mir-125a or mir-125b. J Biol Chem. 2007;282:1479–86.
Akhavantabasi S, Sapmaz A, Tuna S, Erson-Bensan AE. Mir-125b targets ARID3B in breast cancer cells. Cell Struct Funct. 2012;37:27–38.
Guan Y, Yao H, Zheng Z, Qiu G, Sun K. Mir-125b targets BCL3 and suppresses ovarian cancer proliferation. Int J Cancer ;2010.
Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, et al. Mir-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011;71:3552–62.
Huang L, Luo J, Cai Q, Pan Q, Zeng H, Guo Z, et al. MicroRNA-125b suppresses the development of bladder cancer by targeting E2f3. Int J Cancer. 2011;128:1758–69.
Ozek NS, Tuna S, Erson-Bensan AE, Severcan F. Characterization of microRNA-125b expression in MCF7 breast cancer cells by ATR-FTIR spectroscopy. Analyst. 2010;135:3094–102.
Kauffmann A, Gentleman R, Huber W. Arrayqualitymetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–6.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80.
Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–75.
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84.
Falcon S, Gentleman R. Using gostats to test gene lists for go term association. Bioinformatics. 2007;23:257–8.
Akhavantabasi S, Akman HB, Sapmaz A, Keller J, Petty EM, Erson AE. Usp32 is an active, membrane-bound ubiquitin protease overexpressed in breast cancers. Mamm Genome. 2010;21:388–97.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time pcr experiments. Clin Chem U S. 2009;55:611–22.
Gur-Dedeoglu B, Konu O, Bozkurt B, Ergul G, Seckin S, Yulug IG. Identification of endogenous reference genes for qrt-pcr analysis in normal matched breast tumor tissues. Oncol Res. 2009;17:353–65.
Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mrna quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett. 2006;28:1601–13.
Wang J, Gu Z, Ni P, Qiao Y, Chen C, Liu X, et al. NF-kappab p50/p65 hetero-dimer mediates differential regulation of CD166/ALCAM expression via interaction with micoRNA-9 after serum deprivation, providing evidence for a novel negative auto-regulatory loop. Nucleic Acids Res. 2011;39:6440–55.
Jannie KM, Stipp CS, Weiner JA. ALCAM regulates motility, invasiveness, and adherens junction formation in uveal melanoma cells. PLoS One. 2012;7:e39330.
Degen WG, van Kempen LC, Gijzen EG, van Groningen JJ, van Kooyk Y, Bloemers HP, et al. MEMD, a new cell adhesion molecule in metastasizing human melanoma cell lines, is identical to ALCAM (activated leukocyte cell adhesion molecule). Am J Pathol. 1998;152:805–13.
Ofori-Acquah SF, King JA. Activated leukocyte cell adhesion molecule: a new paradox in cancer. Transl Res. 2008;151:122–8.
Burandt E, Bari Noubar T, Lebeau A, Minner S, Burdelski C, Jänicke F, Müller V, Terracciano L, Simon R, Sauter G, Wilczak W, Lebok P. Loss of ALCAM expression is linked to adverse phenotype and poor prognosis in breast cancer: a TMA-based immunohistochemical study on 2,197 breast cancer patients. Oncol Rep. 2014.
Bowen MA, Aruffo AA, Bajorath J. Cell surface receptors and their ligands: in vitro analysis of CD6-CD166 interactions. Proteins. 2000;40:420–8.
Swart GW. Activated leukocyte cell adhesion molecule (CD166/ALCAM): developmental and mechanistic aspects of cell clustering and cell migration. Eur J Cell Biol. 2002;81:313–21.
Weidle UH, Eggle D, Klostermann S, Swart GW. ALCAM/CD166: cancer-related issues. Cancer Genomics Proteomics. 2010;7:231–43.
Graham K, de las Morenas A, Tripathi A, King C, Kavanah M, Mendez J, et al. Gene expression in histologically normal epithelium from breast cancer patients and from cancer-free prophylactic mastectomy patients shares a similar profile. Br J Cancer. 2010;102:1284–93.
Jezierska A, Olszewski WP, Pietruszkiewicz J, Olszewski W, Matysiak W, Motyl T. Activated leukocyte cell adhesion molecule (ALCAM) is associated with suppression of breast cancer cells invasion. Med Sci Monit. 2006;12:BR245–56.
Tan F, Mosunjac M, Adams AL, Adade B, Taye O, Hu Y, et al. Enhanced down-regulation of ALCAM/CD166 in African-American breast cancer. BMC Cancer. 2014;14:715.
Burkhardt M, Mayordomo E, Winzer KJ, Fritzsche F, Gansukh T, Pahl S, et al. Cytoplasmic overexpression of ALCAM is prognostic of disease progression in breast cancer. J Clin Pathol. 2006;59:403–9.
Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–13.
Dusek RL, Bascom JL, Vogel H, Baron S, Borowsky AD, Bissell MJ, et al. Deficiency of the P53/P63 target PERP alters mammary gland homeostasis and promotes cancer. Breast Cancer Res. 2012;14:R65.
Jacobson A, Cunningham JL. Connective tissue growth factor in tumor pathogenesis. Fibrogenesis Tissue Repair. 2012;5 Suppl 1:S8.
Acknowledgments
We would like to thank Dr. Q. Fan and Prof. Ofori-Acquah for sharing ALCAM promoter constructs, Dr. J. Weiner for ALCAM shRNA construct, and Serkan Tuna and Cansaran Saygılı for technical assistance. CS, MTA, and DS were supported by funding from Science Foundation Ireland, IRCSET, and Cancer Research Ireland. This research was funded by METU internal funds and partially by TUBITAK 108S381.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supp. Table 1
Breast cancer patient panel information. (PDF 126 kb)
Supp. Table 2, 3
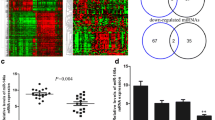
Significantly up-regulated (n=68) and down-regulated genes (n=70) in MCF7-125b cells. (PDF 265 kb)
Supp. Fig. 1
Volcano plot represents the relationship between fold change and statistical significance. X-axis represents the log2 Fold Change between MCF7-EV and MCF7-miR-125b cells. The vertical axis represents the Log Odds (B-Statistic). (PDF 810 kb)
Supp. Fig. 2
A. T47D cells stably transfected with miR-125b or with EV (empty vector, pSUPER). miR-125b expression levels were normalized to U6 expression. Two independent experiments were performed with triplicate replicates. The Student t-test was used for analyzing the data. Transfected cells were polyclonally selected. B. Cell proliferation assay (MTT) was performed in T47D cells, transfected with precursor miR-125b and Empty Vector (EV) as control. Experiment was performed with 4 replicates for each sample. All OD570 values were divided into that of day 1 for both miR-125b and EV cells. 10,000 cells were initially seeded into each well of 96-well cell culture plates and four replicates were used for each sample. C. Seven fold induction of ALCAM expression in MCF7-EV and MCF7-125b cells. Bands were quantified using ImageJ. D. ALCAM protein levels were detectable in T47D-EV cells and no apparent increase was detected upon miR-125b expression. (PDF 256 kb)
Rights and permissions
About this article
Cite this article
Akman, H.B., Selcuklu, S.D., Donoghue, M.T.A. et al. ALCAM is indirectly modulated by miR-125b in MCF7 cells. Tumor Biol. 36, 3511–3520 (2015). https://doi.org/10.1007/s13277-014-2987-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2987-5