Abstract
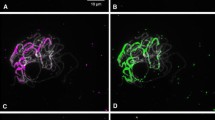
The chromosomal locations of a new class of Revolver transposon-like elements were analyzed by using FISH method on the metaphase chromosome in somatic cell division of the rye cultivar Petkus. First, the Revolver standard element probe λ2 was weakly hybridized throughout the rye chromosome, and comparatively large interstitial signals spotted with a dot shape were detected together with several telomeric regions. The dot shape interstitial signal was stably detected at one site on Chromosome (Chr) 1R (middle part of the interstitial region of the short arm), three sites on Chr 2R (distal part of the interstitial region and adjacent to the centromere on the short arm, middle part of the interstitial region of the long arm), and two sites on Chr 5R (middle part of the interstitial region and adjacent to the centromere on the long arm). The Revolver λ2 probe was effective for identification of 1R, 2R, and 5R chromosomes. On the other hand, Revolver nonautonomous element-specific L626-BARE-100 probe was strongly distributed throughout the rye chromosomes, and considerable numbers and diverse lengths of transcripts were detected by RT-PCR. Although the standard elements were found in localized clusters, the nonautonomous elements tended to be dispersed throughout the genome. Clustered nature of Revolver is a significantly rare case in genomics.





Similar content being viewed by others
References
Altinkut A, Kotseruba V, Kirzhner VH, Nevo E et al (2006) Ac-like transposons in populations of wild diploid Triticeae species: comparative analysis of chromosomal distribution. Chromosome Res 14:307–317
Assis R, Kondrashov AS (2009) Rapid repetitive element-mediated expansion of piRNA clusters in mammalian evolution. Proc Natl Acad Sci USA 106:7079–7082
Barakat A, Carels N, Bernardi G (1997) The distribution of genes in the genomes of Gramineae. Proc Natl Acad Sci USA 94:6857–6861
Bedbrook JR, Jones J, O’Dell M, Thompson RD et al (1980) A molecular description of telomeric heterochromatin in Secale species. Cell 19:545–560
Bennetzen JL (1996) The Mutator transposable element system of maize. Curr Top Microbiol Immunol 204:195–229
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42:251–269
Bennetzen JL (2005) Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev 15:621–627
Bennetzen JL, Wang H (2014) The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol 65:505–530
Bennetzen JL, Wang X (2018) Relationships between gene structure and genome instability in flowering plants. 11:407–413
Bureau TE, Ronald PC, Wessler SR (1996) A computer-based systematic survey reveals the predominance of small inverted- repeat elements in wild-type rice genes. Proc Natl Acad Sci USA 93:8524–8529
Cho J (2018) Transposon-derived non-coding RNAs and their function in plants. Front Plant Sci 9:600
Costa FF (2008) Non-coding RNAs, epigenetics and complexity. Gene 410:9–17
Devor EJ, Huang L, Wise A, Peek AS et al (2011) An X chromosome microRNA cluster in the marsupial species Monodelphis domestica. J Hered 102:577–583
Feschotte C (2008) Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9:397–405
Feschotte C, Jiang N, Wessler SR (2002a) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341
Feschotte C, Zhang X, Wessler SR (2002b) Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. American Society for Microbiology Press, Washington, DC, pp 1147–1158
Flavell RB, Rimpau JR, Smith DB (1977) Repeated sequence DNA relationships in four cereal genomes. Chromosoma 63:205–222
Flavell AJ, Dunbar E, Anderson R, Pearce SR et al (1992) Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res 20:3639–3644
Gill BS, Friebe B, Endo TR (1991) Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34:830–839
Grandbastien MA (1992) Retroelements in higher plants. Trends Genet 8:103–108
Hickman AB, Dyda F (2015) Mechanisms of DNA transposition. Microbiol Spectr 3:MDNA3-0034-2014
Huang CRL, Burns KH, Boeke JD (2012) Active transposition in genomes. Annu Rev Genet 46:51–675
Kalendar R, Tanskanen J, Immonen M, Nevo E et al (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97:6603–6607
Kalendar R, Amenov A, Daniyarov A (2019) Use of retrotransposon-derived genetic markers to analyse genomic variability in plants. Funct Plant Biol 46:15–29
Kempken F, Windhofer F (2001) The hATfamily: a versatile transposon group common to plants, fungi, animals, and man. Chromosoma 110:1–9
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kumar A, Hirochika H (2001) Applications of retrotransposons as genetic tools in plant biology. Trends Plant Sci 6:127–134
Kunze R, Well CF (2002) The hAT and CACTA superfamilies of plant transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. American Society for Microbiology Press, Washington, DC, pp 565–610
Lazarow K, Doll ML, Kunze R (2013) Molecular biology of maize Ac/Ds elements: an overview. Plant Transposable Elements: Methods and Protocols. Methods Mol Biol 1057:59–82
Lee JK, Park JY, Kim JH, Kwon SJ et al (2006) Genetic mapping of the Isaac-CACTA transposon in maize. Theor Appl Genet 113:16–22
Lisch D (2002) Mutator transposons. Trends Plant Sci 7:498–504
Lisch D (2009) Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol 60:43–66
Lisch D (2015) Mutator and MULE transposons. Microbiol Spectr 3:MDNA3-0032-2014
Locke J, Howard LT, Aippersbach N, Podemski L et al (1999) The characterization of DINE-1, a short, interspersed repetitive element present on chromosome and in the centric heterochromatin of Drosophila melanogaster. Chromosoma 108:356–366
Lonnig WE, Saedler H (2012) Chromosome rearrangements and transposable elements. Annu Rev Genet 36:389–410
Lu C, Chen J, Zhang Y et al (2012) Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol 9:1005–1017
Lunde CF, Morrow DJ, Roy LM, Walbot V (2003) Progress in maize gene discovery: a project update. Funct Integr Genomics 3:25–32
Michalak P (2006) RNA world - the dark matter of evolutionary genomics. J Evol Biol19:1768-1774
Muotri AR, Marchetto MCN, Coufal NG et al (2007) The necessary junk: new functions for transposable elements. Hum Mol Genet 16:R159–R167
Myers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11:1660–1676
Nevo E, Beiles A (1989) Genetic diversity of wild emmer wheat in Israel and Turkey. Theor Appl Genet 77:421–455
Nevo E, Korol AB, Beiles A, Fahima T (2002) Evolution of wild emmer and wheat improvement. Population genetics, genetic resources, and genome organization of wheat’s progenitor, Triticum dicoccoides. Springer, Berlin, p 364
Oliver KR, McComb JA, Greene WK (2013) Transposable elements: powerful contributors to angiosperm evolution and diversity. Genome Biol Evol 5:1886–1901
Panstruga R, Buschges R, Piffanelli P, Schulze-Lefert P (1998) A contiguous 60 kb genomic stretch from barley reveals molecular evidence for gene islands in a monocot genome. Nucleic Acids Res 26:1056–1062
Peng J, Ronin YI, Fashima T, Roder MS et al (2003) Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc Natl Acad Sci USA 100:2489–2494
Ramachandran S, Sundaresan V (2001) Transposons as tools for functional genomics. Plant Physiol Biochem 39:243–252
SanMiguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Annu Bot 82:37–44
Siomi H, Siomi MC (2008) Interactions between transposable elements and Argonautes have (probably) been shaping the Drosophila genome throughout evolution. Curr Opin Genet Dev 18:181–187
Song X, Cao X (2017) Transposon-mediated epigenetic regulation contributes to phenotypic diversity and environmental adaptation in rice. Curr Opin Plant Biol 36:111–118
Stein N (2007) Triticeae genomics: advances in sequence analysis of large genome cereal crops. Chromosome Res 15:21–31
Sunker R, Girke T, Zhu JK (2005) Identification and characterization of endogenous small interfering RNAs from rice. Nucleic Acids Res 33:4443–4454
Suoniemi A, Anamthawat-Jonsson K, Arna T, Schulman AH (1996) Retrotransposon BARE-1 is a major, dispersed component of the barley (Hordeum vulgare L.) genome. Plant Mol Biol 30:1321–1329
Tomita M (2008) Revolver-2: a novel transposon-like element from rye. United States Patent 7, 351, 536B2
Tomita M, Shinohara K, Morimoto M (2008) Revolver is a new class of transposon-like gene composing the Triticeae genome. DNA Res 15:49–62
Tomita M, Akai K, Morimoto T (2009) Genomic subtraction recovers rye-specific DNA elements enriched in the rye genome. Mol Biotechnol 42:160–167
Tomita M, Okutani A, Beiles A, Nevo E (2011) Genomic, RNA, and ecological divergences of the Revolver transposon-like multi-gene family in Triticeae. BMC Evol Biol 11 Article number:269
Vicient CM, Suoniemi A, Anamthawat-Jonsson K, Tanskanena J et al (1999) Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11:1769–1784
Vicient CM, Jaaskelainen MJ, Kalendar R, Schulman AH (2001) Active retrotransposons are a common feature of grass genomes. Plant Physiol 125:1283–1129
Wicker T, Sabot F, Hua-Van A, Bennetzen JF et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Wicker T, Narechania A, Sabot F, Stein J et al (2008) Low-pass shotgun sequencing of the barley genome facilitates rapid identification of genes, conserved non-coding sequences and novel repeats. BMC Genomics 9:518
Yuan WY, Tomita M, Sun SC, Yasumuro Y (1998) Identification of multi-alien genome carrying different powdery mildew resistant genes from rye and Haynaldia villosa into wheat genome. Genes Genet Syst 73:377–384
Zhang X, Jiang N, Feschotte C, Wessler SR (2004) PIF- and Pong-like transposable elements: distribution, evolution and relationship with Tourist-like miniature inverted-repeat transposable elements. Genetics 166:971–986
Zhang L, Chen JG, Zhao Q (2015) Regulatory roles of Alu transcript on gene expression. Exp Cell Res 338:113–118
Funding
This work was funded by the Grant-in-Aid for Scientific Research No. 1360006 and No. 04760006 of the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) to Motonori Tomita.
Author information
Authors and Affiliations
Contributions
MT conceived and designed the study; MT, TK, and ET conducted the experiments and analyzed the data; MT wrote the manuscript; all authors read and approved submission.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by: Izabela Pawłowicz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tomita, M., Kanzaki, T. & Tanaka, E. Clustered and dispersed chromosomal distribution of the two classes of Revolver transposon family in rye (Secale cereale). J Appl Genetics 62, 365–372 (2021). https://doi.org/10.1007/s13353-021-00617-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-021-00617-4